2R6W
 
 | | Estrogen receptor alpha ligand-binding domain complexed to a SERM | | Descriptor: | Estrogen receptor, [6-hydroxy-2-(4-hydroxyphenyl)-1-benzothien-3-yl]{4-[2-(4-methylpiperidin-1-yl)ethoxy]phenyl}methanone | | Authors: | Wang, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3B45
 
 | | Crystal structure of GlpG at 1.9A resolution | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
5JFP
 
 | | HIV-1 wild Type protease with GRL-097-13A (a Adamantane P1-Ligand with bis-THF in P2 and isobutylamine in P1') | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
5JFU
 
 | | HIV-1 wild Type protease with GRL-007-14A (a Adamantane P1-Ligand with bis-THF in P2 and benzylamine in P1') | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-{benzyl[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
2R6Y
 
 | | Estrogen receptor alpha ligand-binding domain in complex with a SERM | | Descriptor: | Estrogen receptor, [6-hydroxy-2-(4-hydroxyphenyl)-1-benzothien-3-yl][4-(2-pyrrolidin-1-ylethoxy)phenyl]methanone | | Authors: | Wang, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7K0N
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5JG1
 
 | | HIV-1 wild Type protease with GRL-031-14A (a Adamantane P1-Ligand with tetrahydropyrano-tetrahydrofuran in P2 and isobutylamine in P1') | | Descriptor: | (3R,3aS,7aR)-hexahydro-4H-furo[2,3-b]pyran-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
7V9X
 
 | |
7E04
 
 | |
5GWI
 
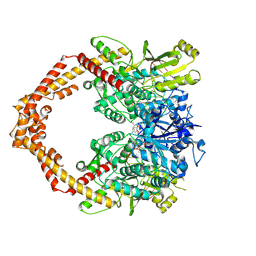 | | Structure of a Human topoisomerase IIbeta fragment in complex with DNA and E7873R | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Wang, Y.R, Chen, S.F, Wu, C.C, Chan, N.L. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.737 Å) | | Cite: | Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry
Nucleic Acids Res., 45, 2017
|
|
5Y03
 
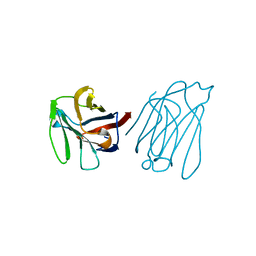 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138.
Sci Rep, 8, 2018
|
|
1D8V
 
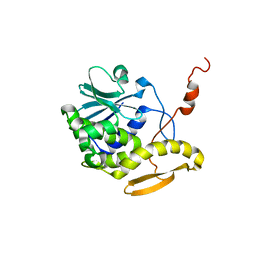 | | THE RESTRAINED AND MINIMIZED AVERAGE NMR STRUCTURE OF MAP30. | | Descriptor: | ANTI-HIV AND ANTI-TUMOR PROTEIN MAP30 | | Authors: | Wang, Y.-X, Neamati, N, Jacob, J, Palmer, I, Stahl, S.J. | | Deposit date: | 1999-10-26 | | Release date: | 1999-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of anti-HIV-1 and anti-tumor protein MAP30: structural insights into its multiple functions.
Cell(Cambridge,Mass.), 99, 1999
|
|
5GWK
 
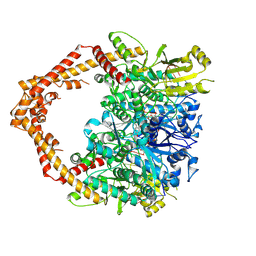 | | Human topoisomerase IIalpha in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wang, Y.R, Wu, C.C, Chan, N.L. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry
Nucleic Acids Res., 45, 2017
|
|
5GSW
 
 | | Crystal structure of EV71 3C in complex with N69S 1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-17 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
5GSO
 
 | | Crystal Structures of EV71 3C Protease in complex with NK-1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
1XTA
 
 | | Crystal Structure of Natrin, a snake venom CRISP from Taiwan cobra (Naja atra) | | Descriptor: | Natrin 1 | | Authors: | Wang, Y.-L, Goh, K.-X, Lee, S.-C, Huang, W.-N, Wu, W.-G, Chen, C.-J. | | Deposit date: | 2004-10-21 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of snake venom CRISP reveal an action mechanism involving serine protease and ion channel blocking domains
To be Published
|
|
6DFX
 
 | | human diabetogenic TCR T1D3 in complex with DQ8-p8E9E peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
5ZED
 
 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (E214V/T215S) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein ADH | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
6DFQ
 
 | | mouse diabetogenic TCR I.29 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TCR alpha chain, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
4J54
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical isolate PR20 with the potent antiviral inhibitor GRL-0519A | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, ACETATE ION, IODIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Extreme Multidrug Resistant HIV-1 Protease with 20 Mutations Is Resistant to Novel Protease Inhibitors with P1'-Pyrrolidinone or P2-Tris-tetrahydrofuran.
J.Med.Chem., 56, 2013
|
|
6DFV
 
 | | Mouse diabetogenic TCR 8F10 | | Descriptor: | 1,2-ETHANEDIOL, TCR alpha chain, TCR beta chain | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
5WQJ
 
 | |
5EX7
 
 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
5WQK
 
 | |
6DFS
 
 | | mouse TCR I.29 in complex with IAg7-p8E9E6ss | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
