8X1V
 
 | |
8HXB
 
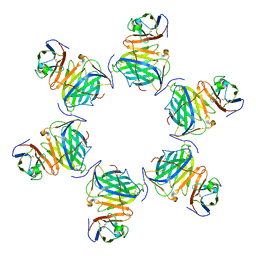 | | Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXC
 
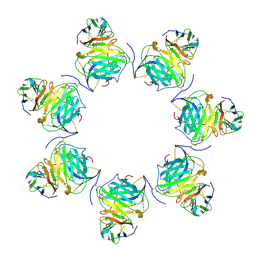 | | Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXA
 
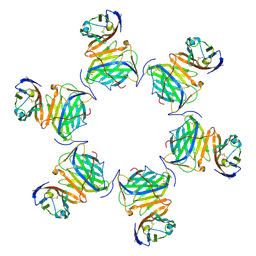 | | Cryo-EM structure of MPXV M2 in complex with human B7.1 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD80 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
6ISO
 
 | | Human SIRT3 Recognizing H3K4cr | | Descriptor: | (2E)-BUT-2-ENAL, ARG-THR-LYS-GLN-THR-ALA-ARG, GLYCEROL, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2018-11-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Elife, 3, 2014
|
|
8ITR
 
 | |
8I7Q
 
 | |
4TVU
 
 | | Crystal structure of trehalose synthase from Deinococcus radiodurans reveals a closed conformation for catalysis of the intramolecular isomerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Liaw, S.H. | | Deposit date: | 2014-06-28 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9IJB
 
 | |
8XAU
 
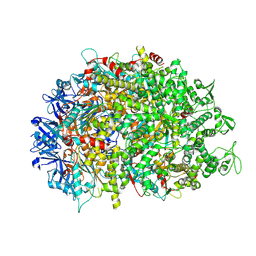 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
6IDY
 
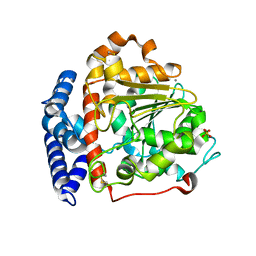 | |
8HVR
 
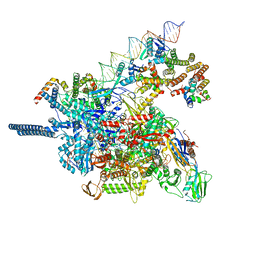 | |
9IID
 
 | | Crystal structure of PgAfp | | Descriptor: | Antifungal protein B | | Authors: | Wang, Y. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of PgAfp
To Be Published
|
|
4WF7
 
 | | Crystal structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in the intramolecular isomerization catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Hsieh, Y.C, Lee, G.C, Liaw, S.H. | | Deposit date: | 2014-09-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4U8W
 
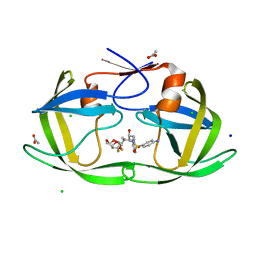 | | HIV-1 wild Type protease with GRL-050-10A (a Gem-difluoro-bis-Tetrahydrofuran as P2-Ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of gem-Difluoro-bis-Tetrahydrofuran as P2 Ligand for HIV-1 Protease Inhibitors to Improve Brain Penetration: Synthesis, X-ray Studies, and Biological Evaluation.
Chemmedchem, 10, 2015
|
|
6X11
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - an enol tautomer of ACMS monodentately bound structure | | Descriptor: | (2Z,3Z)-2-[(2Z)-3-hydroxyprop-2-en-1-ylidene]-3-iminobutanedioic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-05-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8AOU
 
 | |
6XJX
 
 | | MCU holocomplex in Low-calcium blocking state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
6XJV
 
 | | MCU holocomplex in High-calcium state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
7WD0
 
 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | |
7WD7
 
 | |
7WCZ
 
 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
 | |
