1IC1
 
 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5O69
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
3NE6
 
 | | RB69 DNA Polymerase (S565G/Y567A) Ternary Complex with dCTP Opposite dG | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-08 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
3NGI
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite dG | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
1G6M
 
 | |
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
2ERF
 
 | |
3JTN
 
 | | Crystal Structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2, IODIDE ION | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the MecA degradation tag
To be Published
|
|
3JTP
 
 | | crystal structure of the C-terminal domain of MecA | | Descriptor: | Adapter protein mecA 1, IODIDE ION | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | crystal structure of the MecA degradation tag
To be Published
|
|
1LSL
 
 | | Crystal Structure of the Thrombospondin-1 Type 1 Repeats | | Descriptor: | Thrombospondin 1, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Tan, K, Duquette, M, Liu, J, Dong, Y, Zhang, R, Joachimiak, A, Lawler, J, Wang, J.-H. | | Deposit date: | 2002-05-17 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the TSP-1 type 1 repeats: a novel
layered fold and its biological implication.
J.Cell Biol., 159, 2002
|
|
2ES3
 
 | |
4B99
 
 | | Crystal Structure of MAPK7 (ERK5) with inhibitor | | Descriptor: | 11-cyclopentyl-2-[[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl-phenyl]amino]-5-methyl-pyrimido[4,5-b][1,4]benzodiazepin-6-one, MITOGEN-ACTIVATED PROTEIN KINASE 7 | | Authors: | Elkins, J.M, Wang, J, Vollmar, M, Mahajan, P, Savitsky, P, Deng, X, Gray, N.S, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-03 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Crystal Structure of Erk5 (Mapk7) in Complex with a Specific Inhibitor.
J.Med.Chem., 56, 2013
|
|
6UIB
 
 | | Crystal structure of IL23 bound to peptide 23-652 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Durbin, J.D, Wang, J, Afshar, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Integration of phage and yeast display platforms: A reliable and cost effective approach for binning of peptides as displayed on-phage.
Plos One, 15, 2020
|
|
1TAU
 
 | | TAQ POLYMERASE (E.C.2.7.7.7)/DNA/B-OCTYLGLUCOSIDE COMPLEX | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, DNA (5'-D(*CP*GP*GP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*CP*CP*G)-3'), ... | | Authors: | Eom, S.H, Wang, J, Steitz, T.A. | | Deposit date: | 1996-06-17 | | Release date: | 1997-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Taq ploymerase with DNA at the polymerase active site.
Nature, 382, 1996
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
4YR8
 
 | | Crystal structure of JNK in complex with a regulator protein | | Descriptor: | CHLORIDE ION, Dual specificity protein phosphatase 16, Mitogen-activated protein kinase 8 | | Authors: | Liu, X, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2015-03-14 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A conserved motif in JNK/p38-specific MAPK phosphatases as a determinant for JNK1 recognition and inactivation.
Nat Commun, 7, 2016
|
|
7MG0
 
 | |
8YAI
 
 | | Crystal structure of glucose 1-dehydrogenase mutant1 from Limosilactobacillus fermentum | | Descriptor: | SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
8YAV
 
 | | Crystal structure of glucose 1-dehydrogenase from Limosilactobacillus fermentum | | Descriptor: | MAGNESIUM ION, SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-10 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir.
Adv.Synth.Catal., n/a, 2024
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
4RNG
 
 | | Crystal structure of a bacterial homologue of SWEET transporters | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MtN3/saliva family, SULFATE ION | | Authors: | Hu, Q, Wang, J, Yan, C, Yan, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a bacterial homologue of SWEET transporters.
Cell Res., 24, 2014
|
|
5UFL
 
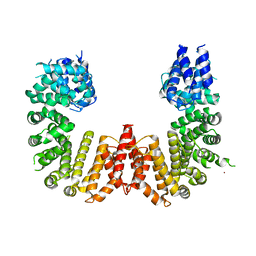 | | Crystal structure of a CIP2A core domain | | Descriptor: | Protein CIP2A, ZINC ION | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oncoprotein CIP2A is stabilized via interaction with tumor suppressor PP2A/B56.
EMBO Rep., 18, 2017
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
8W8R
 
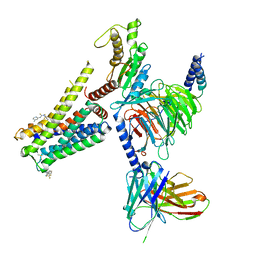 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
