6TYD
 
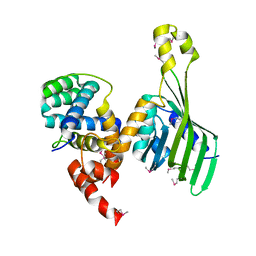 | | Structure of human LDB1 in complex with SSBP2 | | Descriptor: | LIM domain-binding protein 1, Single-stranded DNA-binding protein 2 | | Authors: | Wang, H, Wang, Z, Xu, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of human LDB1 in complex with SSBP2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5YQL
 
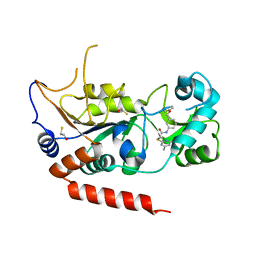 | | Crystal structure of Sirt2 in complex with selective inhibitor A2I | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[3-(phenoxymethyl)phenyl]ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
4QBR
 
 | |
8JHO
 
 | |
1YNS
 
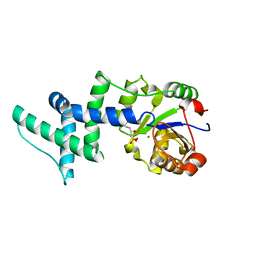 | | Crystal Structure Of Human Enolase-phosphatase E1 and its complex with a substrate analog | | Descriptor: | 2-OXOHEPTYLPHOSPHONIC ACID, E-1 enzyme, MAGNESIUM ION | | Authors: | Wang, H, Pang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human e1 enzyme and its complex with a substrate analog reveals the mechanism of its phosphatase/enolase
J.Mol.Biol., 348, 2005
|
|
8X8P
 
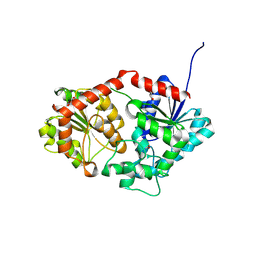 | |
6K8N
 
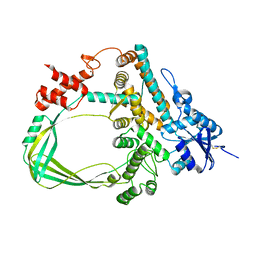 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III | | Descriptor: | ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
1ZS9
 
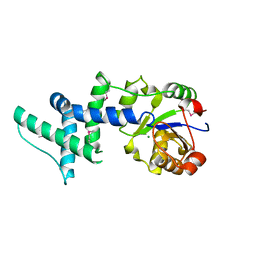 | | Crystal structure of human enolase-phosphatase E1 | | Descriptor: | E-1 ENZYME, MAGNESIUM ION | | Authors: | Wang, H, Pang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-23 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human E1 Enzyme and its Complex with a Substrate Analog Reveals the Mechanism of its Phosphatase/Enolase
J.Mol.Biol., 348, 2005
|
|
2OUU
 
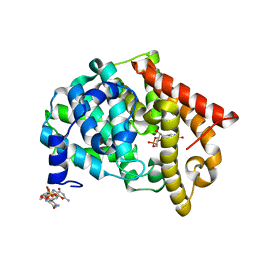 | | crystal structure of PDE10A2 mutant D674A in complex with cGMP | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4MMC
 
 | |
1WWI
 
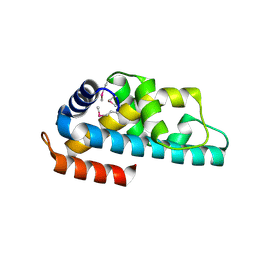 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
2BPR
 
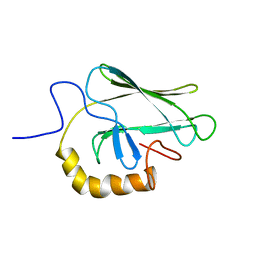 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, 25 STRUCTURES | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
1RMV
 
 | | RIBGRASS MOSAIC VIRUS, FIBER DIFFRACTION | | Descriptor: | RIBGRASS MOSAIC VIRUS COAT PROTEIN, RIBGRASS MOSAIC VIRUS RNA | | Authors: | Wang, H, Stubbs, G. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | FIBER DIFFRACTION (2.9 Å) | | Cite: | Molecular dynamics in refinement against fiber diffraction data.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
3O47
 
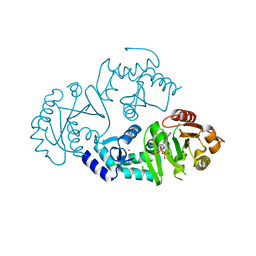 | | Crystal structure of ARFGAP1-ARF1 fusion protein | | Descriptor: | ADP-ribosylation factor GTPase-activating protein 1, ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Tong, Y, Nedyalkova, L, Tempel, W, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ARFGAP1-ARF1 fusion protein
to be published
|
|
3MF0
 
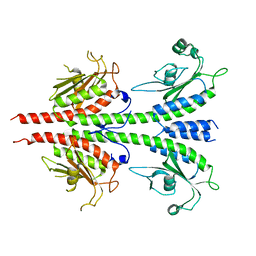 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
4MM8
 
 | |
4MM9
 
 | |
8IQK
 
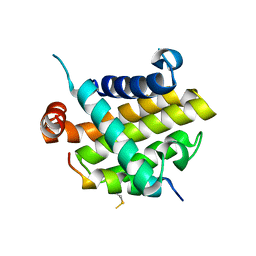 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl-2-like protein 1, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
4MM5
 
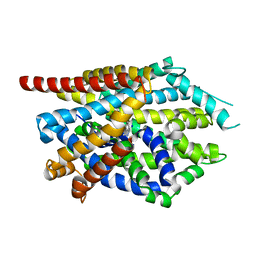 | | Crystal structure of LeuBAT (delta13 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMB
 
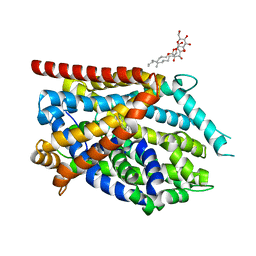 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
8IKR
 
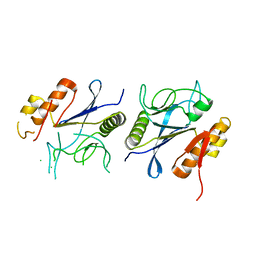 | | Crystal structure of DpaA | | Descriptor: | YkuD domain-containing protein | | Authors: | Wang, H.-J, Hsieh, K.-Y, Lee, S.-H, Chang, C.-I. | | Deposit date: | 2023-03-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the hydrolytic activity of the transpeptidase-like protein DpaA to detach Braun's lipoprotein from peptidoglycan.
Mbio, 14, 2023
|
|
2OUR
 
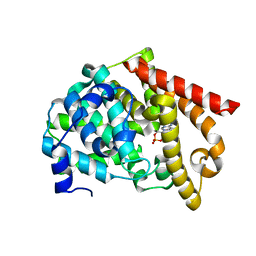 | | crystal structure of PDE10A2 mutant D674A in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6BYF
 
 | |
4ZYO
 
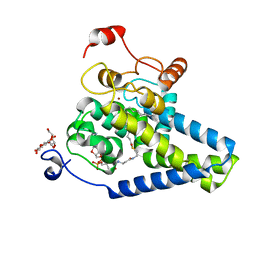 | | Crystal Structure of Human Integral Membrane Stearoyl-CoA Desaturase with Substrate | | Descriptor: | Acyl-CoA desaturase, DODECYL-BETA-D-MALTOSIDE, STEAROYL-COENZYME A, ... | | Authors: | Wang, H, Klein, M.G, Lane, W, Snell, G, Levin, I, Li, K, Zou, H, Sang, B.-C. | | Deposit date: | 2015-05-21 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2PW3
 
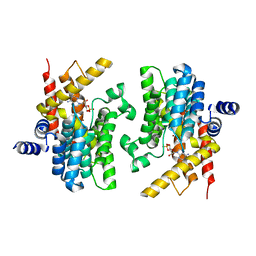 | | Structure of the PDE4D-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The molecular basis for different recognition of substrates by phosphodiesterase families 4 and 10.
J.Mol.Biol., 371, 2007
|
|
