4LN0
 
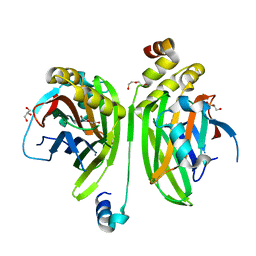 | | Crystal structure of the VGLL4-TEAD4 complex | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Transcription cofactor vestigial-like protein 4, ... | | 著者 | Wang, H, Shi, Z, Zhou, Z. | | 登録日 | 2013-07-11 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.896 Å) | | 主引用文献 | A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer.
Cancer Cell, 25, 2014
|
|
6M8C
 
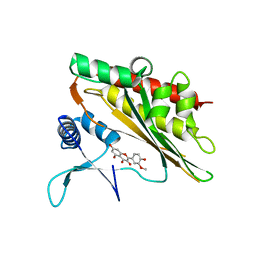 | |
6M8E
 
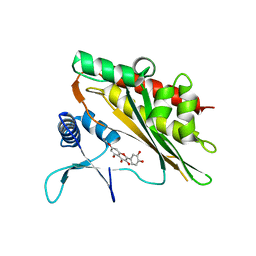 | |
1FBN
 
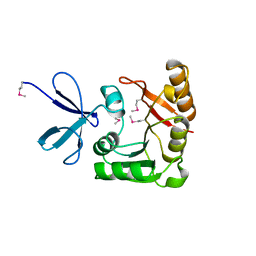 | | CRYSTAL STRUCTURE OF A FIBRILLARIN HOMOLOGUE FROM METHANOCOCCUS JANNASCHII, A HYPERTHERMOPHILE, AT 1.6 A | | 分子名称: | MJ FIBRILLARIN HOMOLOGUE | | 著者 | Wang, H, Boisvert, D, Kim, K.K, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 1999-04-25 | | 公開日 | 2000-04-26 | | 最終更新日 | 2014-11-26 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution.
EMBO J., 19, 2000
|
|
6M8A
 
 | |
6VCD
 
 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | 分子名称: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | 著者 | Wang, H, Shi, H, Zheng, N. | | 登録日 | 2019-12-20 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
8I0E
 
 | | Sb3GT1 complex with UDP | | 分子名称: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Wang, H.T, Wang, Z.L, Ye, M. | | 登録日 | 2023-01-10 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
8I0D
 
 | |
6M88
 
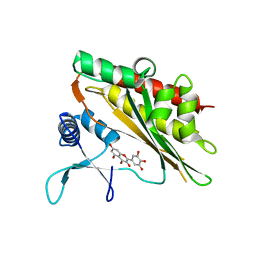 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with myricetin | | 分子名称: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2018-08-21 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Inhibition of Inositol Polyphosphate Kinases by Quercetin and Related Flavonoids: A Structure-Activity Analysis.
J. Med. Chem., 62, 2019
|
|
1ZKL
 
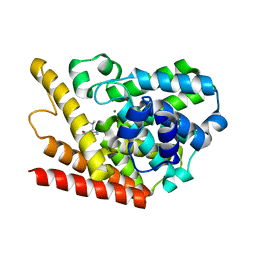 | | Multiple Determinants for Inhibitor Selectivity of Cyclic Nucleotide Phosphodiesterases | | 分子名称: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | 著者 | Wang, H, Liu, Y, Chen, Y, Robinson, H, Ke, H. | | 登録日 | 2005-05-03 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7
J.Biol.Chem., 280, 2005
|
|
6KA0
 
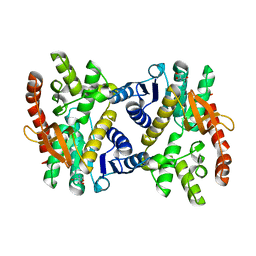 | |
6KA1
 
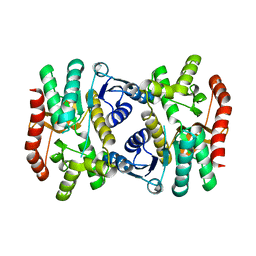 | |
1V8Q
 
 | | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8 | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TT0826 | | 著者 | Wang, H, Takemoto-Hori, C, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-01-13 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
5ZM8
 
 | | Crystal structure of ORP2-ORD in complex with PI(4,5)P2 | | 分子名称: | Oxysterol-binding protein-related protein 2, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | 著者 | Wang, H, Dong, J.Q, Wang, J, Wu, J.W. | | 登録日 | 2018-04-01 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | ORP2 Delivers Cholesterol to the Plasma Membrane in Exchange for Phosphatidylinositol 4, 5-Bisphosphate (PI(4,5)P2).
Mol. Cell, 73, 2019
|
|
6A9S
 
 | | The crystal structure of vaccinia virus A26 (residues 1-397) | | 分子名称: | 1,2-ETHANEDIOL, Protein A26 | | 著者 | Wang, H.C, Ko, T.Z, Luo, Y.C, Liao, Y.T, Chang, W. | | 登録日 | 2018-07-16 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Vaccinia viral A26 protein is a fusion suppressor of mature virus and triggers membrane fusion through conformational change at low pH.
Plos Pathog., 15, 2019
|
|
6LZH
 
 | | Crystal structure of Alpha/beta hydrolase GrgF from Penicillium sp. sh18 | | 分子名称: | GrgF, SODIUM ION | | 著者 | Wang, H, Yu, J, Wang, W.G, Matsuda, Y, Yao, M. | | 登録日 | 2020-02-19 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Basis for the Biosynthesis of an Unusual Chain-Fused Polyketide, Gregatin A.
J.Am.Chem.Soc., 142, 2020
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | 著者 | Wang, H, Liu, X, Sun, L, Yang, H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
3WOH
 
 | |
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | 分子名称: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | 著者 | Wang, H, Shao, M, Sun, L, Yang, H. | | 登録日 | 2022-11-12 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
4MM4
 
 | |
4MMF
 
 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | 分子名称: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | 著者 | Wang, H, Gouaux, E. | | 登録日 | 2013-09-08 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4RGW
 
 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | 分子名称: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | 著者 | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | 登録日 | 2014-09-30 | | 公開日 | 2014-12-03 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
5YQM
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A29 | | 分子名称: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-(4-phenylsulfanylphenyl)ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | 著者 | Wang, H, Yu, Y, Li, G, chen, Q. | | 登録日 | 2017-11-07 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.735 Å) | | 主引用文献 | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
4MMD
 
 | |
5Z3W
 
 | | Malate dehydrogenase binds silver at C113 | | 分子名称: | Malate dehydrogenase, SILVER ION | | 著者 | Wang, H, Wang, M, Sun, H. | | 登録日 | 2018-01-09 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm
Chem Sci, 2020
|
|
