7WKU
 
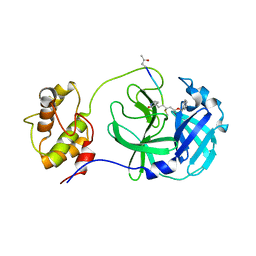 | | Structure of PDCoV Mpro in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Peptidase C30 | | Authors: | Wang, F.H, Yang, H.T. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.
Viruses, 14, 2022
|
|
6D5F
 
 | | Cryo-EM reconstruction of membrane-enveloped filamentous virus SFV1 (Sulfolobus filamentous virus 1) | | Descriptor: | DNA (336-MER), Fimbrial protein | | Authors: | Wang, F, Osinski, T, Liu, Y, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-08-29 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile.
Nat Commun, 9, 2018
|
|
3FNH
 
 | | Crystal structure of InhA bound to triclosan derivative | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(2-PHENYLETHYL)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F. | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Triclosan Derivatives: Towards Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Chemmedchem, 4, 2009
|
|
6JWE
 
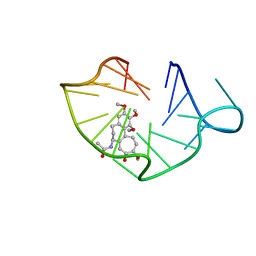 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
3FNG
 
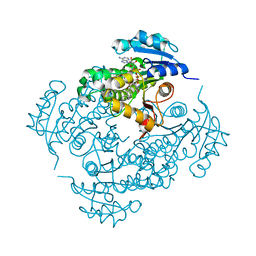 | | Crystal structure of InhA bound to triclosan derivative | | Descriptor: | 5-(cyclohexylmethyl)-2-(2,4-dichlorophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F. | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Triclosan Derivatives: Towards Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Chemmedchem, 4, 2009
|
|
4PIW
 
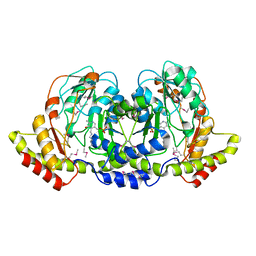 | | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12 | | Descriptor: | TDP-4-keto-6-deoxy-D-glucose transaminase family protein | | Authors: | Wang, F, Xu, W, Helmich, K.E, Singh, S, Yennamalli, R.M, Miller, M.D, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12
To Be Published
|
|
4K0B
 
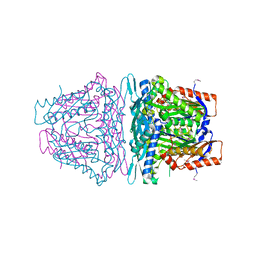 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
7KHD
 
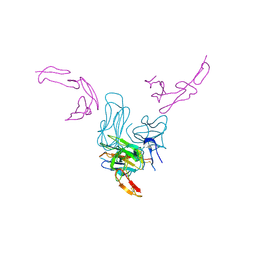 | | Human GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.956102 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
7KHX
 
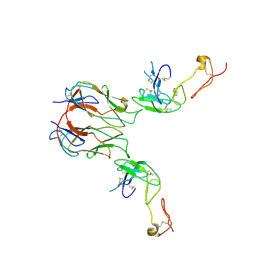 | | Mouse GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, ... | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.20569849 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
8WF7
 
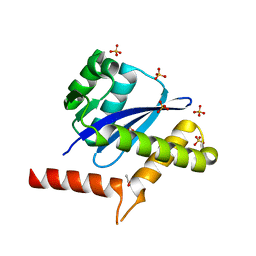 | | The Crystal Structure of integrase from Biortus | | Descriptor: | ACETATE ION, Integrase, SULFATE ION | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of integrase from Biortus
To Be Published
|
|
8WD2
 
 | | The Crystal Structure of p53 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Lu, Y. | | Deposit date: | 2023-09-14 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of p53 from Biortus.
To Be Published
|
|
8WF4
 
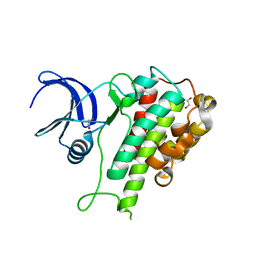 | | The Crystal Structure of RSK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of RSK1 from Biortus.
To Be Published
|
|
8WFY
 
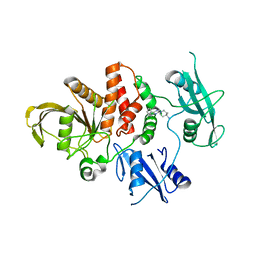 | | The Crystal Structure of SHP2 from Biortus. | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of SHP2 from Biortus.
To Be Published
|
|
8WD3
 
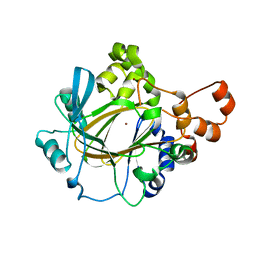 | | The Crystal Structure of JMJD2A(M1-L359) from Biortus. | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2023-09-14 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of JMJD2A(M1-L359) from Biortus.
To Be Published
|
|
8WFQ
 
 | | The Crystal Structure of RALDH1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase 1A1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Shen, Z. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of RALDH1 from Biortus.
To Be Published
|
|
8WFE
 
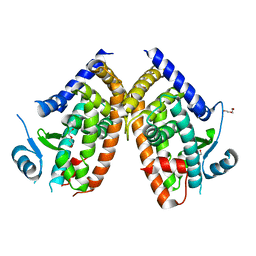 | | The Crystal Structure of PPARg from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, F, Cheng, W, Lv, Z, Guo, S, Lin, D. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of PPARg from Biortus.
To Be Published
|
|
8WFR
 
 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
8XOY
 
 | | The Crystal Structure of PTP1B from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Li, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of PTP1B from Biortus.
To Be Published
|
|
8WSW
 
 | | The Crystal Structure of LIMK2a from Biortus | | Descriptor: | 1,2-ETHANEDIOL, LIM domain kinase 2, ~{N}-[5-[2-[2,6-bis(chloranyl)phenyl]-5-[bis(fluoranyl)methyl]pyrazol-3-yl]-1,3-thiazol-2-yl]-2-methyl-propanamide | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of LIMK2a from Biortus.
To Be Published
|
|
8WTF
 
 | | The Crystal Structure of IRAK4 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of IRAK4 from Biortus
To Be Published
|
|
8X2S
 
 | | The Crystal Structure of BPGM from Biortus | | Descriptor: | 1,2-ETHANEDIOL, Bisphosphoglycerate mutase | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of BPGM from Biortus
To Be Published
|
|
8X2R
 
 | | The Crystal Structure of HSP 90-alpha from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of HSP 90-alpha from Biortus.
To Be Published
|
|
8WUG
 
 | | The Crystal Structure of JMJD2D from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lysine-specific demethylase 4D, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2023-10-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of JMJD2D from Biortus.
To Be Published
|
|
