6BZE
 
 | | Cryo-EM structure of BCL10 CARD filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Li, Y, Ma, J, Garner, E, Zhang, X, Wu, H. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3SFW
 
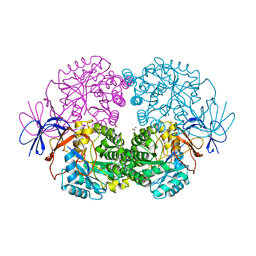 | |
6MK7
 
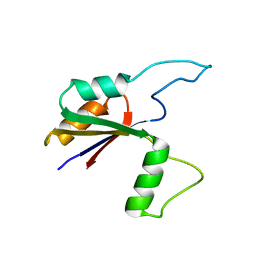 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | Descriptor: | Cell division protein FtsX | | Authors: | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | Deposit date: | 2018-09-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
2M21
 
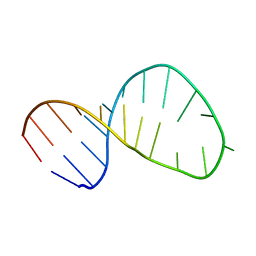 | | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*UP*AP*CP*AP*CP*UP*AP*UP*UP*UP*AP*UP*CP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Wu, H, Trantirek, L, O'Connor, C.M, Feigon, J, Collins, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Rna, 12, 2006
|
|
2KBK
 
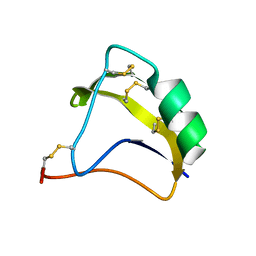 | | Solution Structure of BmK-M10 | | Descriptor: | Neurotoxin BmK-M10 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmK-M10
To be Published
|
|
2NA6
 
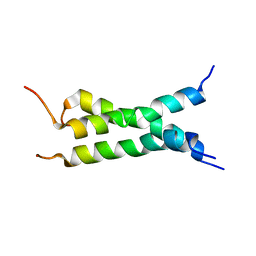 | | Transmembrane domain of mouse Fas/CD95 death receptor | | Descriptor: | Tumor necrosis factor receptor superfamily member 6 | | Authors: | Fu, Q, Chou, J.J, Wu, H, Fu, T, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor.
Mol.Cell, 61, 2016
|
|
5WCF
 
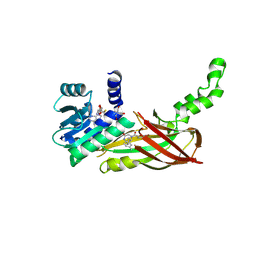 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | (5R)-4-(5-bromofuran-2-carbonyl)-5-(4-fluorophenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Zeng, H, Hutch, A, Seitova, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with MTLLE1441
to be published
|
|
9BOL
 
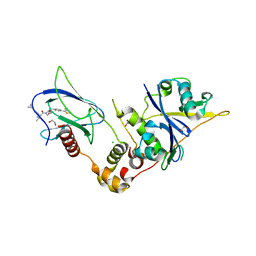 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | (4R)-1-[(2S)-2-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-3,3-dimethylbutanoyl]-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potency-Enhanced Peptidomimetic VHL Ligands with Improved Oral Bioavailability.
J.Med.Chem., 67, 2024
|
|
6MIZ
 
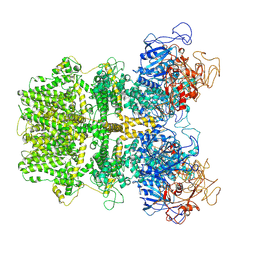 | |
5WCI
 
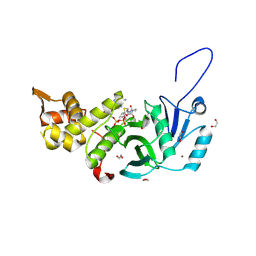 | | Human MYST histone acetyltransferase 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Zheng, Y.G, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Human MYST histone acetyltransferase 1
to be published
|
|
6MIX
 
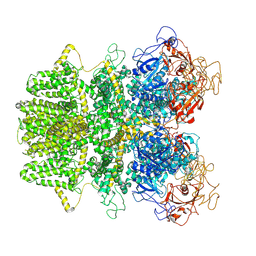 | | Human TRPM2 ion channel in apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
2K1Z
 
 | |
2NS5
 
 | |
2OGP
 
 | | Solution structure of the second PDZ domain of Par-3 | | Descriptor: | Partitioning-defective 3 homolog | | Authors: | Feng, W, Wu, H, Chen, J, Chan, L.-N, Zhang, M. | | Deposit date: | 2007-01-07 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | PDZ domains of par-3 as potential phosphoinositide signaling integrators
Mol.Cell, 28, 2007
|
|
2PON
 
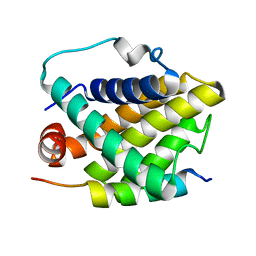 | | Solution structure of the Bcl-xL/Beclin-1 complex | | Descriptor: | Apoptosis regulator Bcl-X, Beclin-1 | | Authors: | Feng, W, Huang, S, Wu, H, Zhang, M. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Bcl-xL's Target Recognition Versatility Revealed by the Structure of Bcl-xL in Complex with the BH3 Domain of Beclin-1.
J.Mol.Biol., 372, 2007
|
|
4LWD
 
 | | Human CARMA1 CARD domain | | Descriptor: | Caspase recruitment domain-containing protein 11, MAGNESIUM ION, SULFATE ION | | Authors: | Zheng, C, Wu, H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
5WCG
 
 | | SET and MYND Domain Containing protein 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, N-lysine methyltransferase SMYD2, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Hutch, A, Seitova, A, Tatlock, J, Kumpf, R, Owen, A, Taylor, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of SMYD2 in complex with compound MTF003
to be published
|
|
2PKU
 
 | | Solution structure of PICK1 PDZ in complex with the carboxyl tail peptide of GluR2 | | Descriptor: | PRKCA-binding protein, peptide (GLU)(SER)(VAL)(LYS)(ILE) | | Authors: | Pan, L, Wu, H, Shen, C, Shi, Y, Xia, J, Zhang, M. | | Deposit date: | 2007-04-18 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes
Embo J., 26, 2007
|
|
3QAK
 
 | | Agonist bound structure of the human adenosine A2a receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,lysozyme chimera | | Authors: | Xu, F, Wu, H, Katritch, V, Han, G.W, Cherezov, V, Stevens, R, GPCR Network (GPCR) | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an agonist-bound human A2A adenosine receptor.
Science, 332, 2011
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
4N0F
 
 | |
2K20
 
 | |
2LIR
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in oxidized states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
