8X5N
 
 | | Structure of ATP/Mg2+ bound Gabija GajA-GajB 4:4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
3Q52
 
 | |
3Q53
 
 | |
6O3Z
 
 | | Crystal structure of RORgt with 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide, RAR-related orphan receptor C isoform a variant | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of [1,2,4]Triazolo[1,5-a]pyridine Derivatives as Potent and Orally Bioavailable ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 11, 2020
|
|
5VT2
 
 | | Crystal structure of growth differentiation factor | | Descriptor: | 1,2-ETHANEDIOL, Growth/differentiation factor 15 | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2017-05-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys.
Sci Transl Med, 9, 2017
|
|
3NK6
 
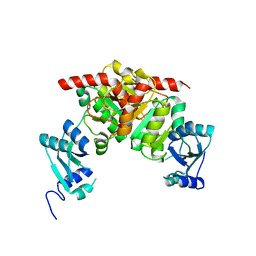 | | Structure of the Nosiheptide-resistance methyltransferase | | Descriptor: | 23S rRNA methyltransferase | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3Q4Z
 
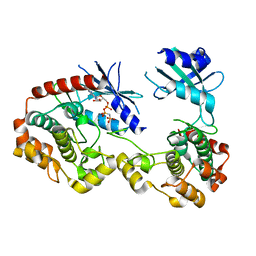 | | Structure of unphosphorylated PAK1 kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 1 | | Authors: | Wang, J, Wu, J.-W, Wang, Z.-X. | | Deposit date: | 2010-12-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural insights into the autoactivation mechanism of p21-activated protein kinase
Structure, 19, 2011
|
|
6DKJ
 
 | | human GIPR ECD and Fab complex | | Descriptor: | 1,2-ETHANEDIOL, Fab heavy chain, Fab light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models.
Sci Transl Med, 10, 2018
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NK7
 
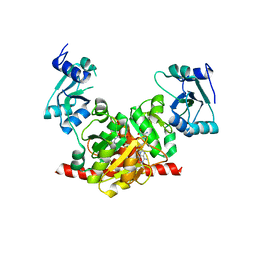 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | Descriptor: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
4LWW
 
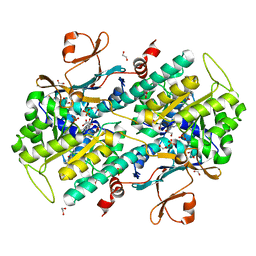 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-(4-(phenylsulfonyl)benzyl)-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7XBR
 
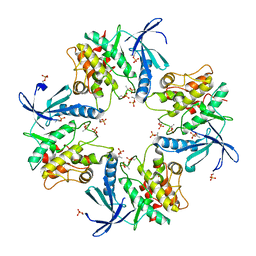 | | Crystal structure of phosphorylated AtMKK5 | | Descriptor: | Mitogen-activated protein kinase kinase 5 | | Authors: | Pei, C.J, Luo, Z.P, Wu, J.W, Wang, Z.X. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the phosphorylated Arabidopsis MKK5 reveals activation mechanism of MAPK kinases.
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7XSC
 
 | |
4RMZ
 
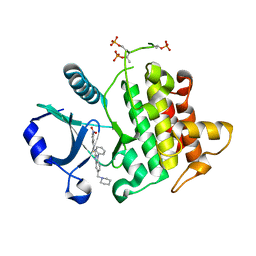 | | Crystal Structure of IRAK-4 | | Descriptor: | 3-nitro-N-[1-phenyl-5-(piperidin-1-ylmethyl)-1H-benzimidazol-2-yl]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Johnstone, S, Sudom, A, Liu, J, Walker, N.P, Wang, Z. | | Deposit date: | 2014-10-22 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of IRAK-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper
To be Published
|
|
7V5K
 
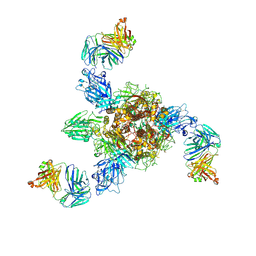 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1)
to be published
|
|
7V5J
 
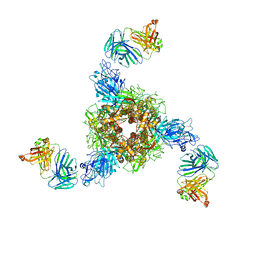 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2)
to be published
|
|
7V6N
 
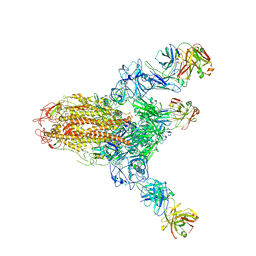 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1 | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1
to be published
|
|
7V6O
 
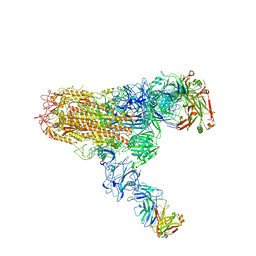 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2) | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2)
to be published
|
|
7V3L
 
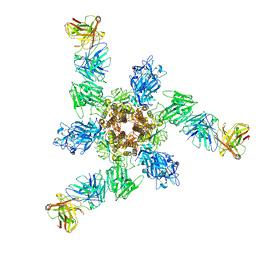 | | MERS S ectodomain trimer in complex with neutralizing antibody 6516 | | Descriptor: | Spike glycoprotein, antibody H, antibody L | | Authors: | Wang, X, Zhao, J, Wang, Z, Wang, Y, Zeng, J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 6516
to be published
|
|
5LZE
 
 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
4LNQ
 
 | |
5LZA
 
 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZF
 
 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
8YZS
 
 | | Structure of the NACC1 BEN domain in complex with its target DNA | | Descriptor: | CATG-containing DNA, Nucleus accumbens-associated protein 1 | | Authors: | Ren, J, Wang, Z. | | Deposit date: | 2024-04-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of DNA recognition by BEN domain proteins reveals a role for oligomerization in unmethylated DNA selection by BANP.
Nucleic Acids Res., 52, 2024
|
|
