8HNV
 
 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | Descriptor: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14182 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7UFG
 
 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
3KCV
 
 | | Structure of formate channel | | Descriptor: | Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
1F03
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
4Z64
 
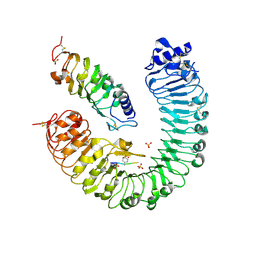 | | the plant peptide hormone receptor complex in arabidopsis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine, ... | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
4Z63
 
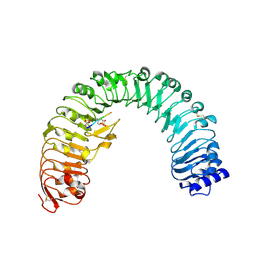 | | The plant peptide hormone receptor in arabidopsis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine, Phytosulfokine receptor 1 | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
4Z5W
 
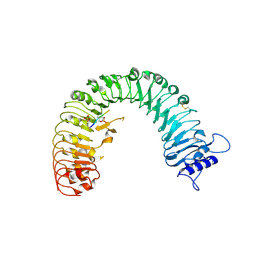 | | The plant peptide hormone receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine, ... | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
5ZAP
 
 | | Atomic structure of the herpes simplex virus type 2 B-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Yuan, S, Wang, J.L, Zhu, D.J, Wang, N, Gao, Q, Chen, W.Y, Tang, H, Wang, J.Z, Zhang, X.Z, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-18 | | Last modified: | 2018-09-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a herpesvirus capsid at 3.1 angstrom.
Science, 360, 2018
|
|
4Z61
 
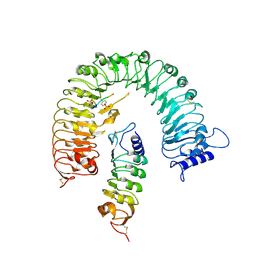 | | The plant peptide hormone receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-ILE-PTR-THR-GLN, Phytosulfokine receptor 1, ... | | Authors: | Chai, J, Wang, J. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
6Z70
 
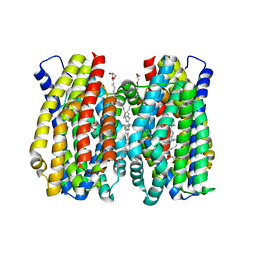 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4CSU
 
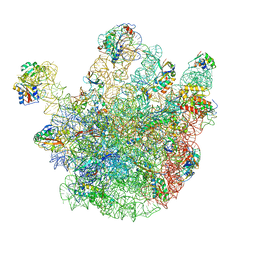 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
6FV7
 
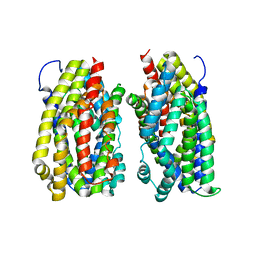 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dimer structure of Aq128 in the outward-facing state
To Be Published
|
|
1I5U
 
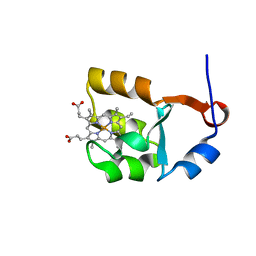 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
4D90
 
 | | Crystal Structure of Del-1 EGF domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Schurpf, T, Springer, T, Wang, J. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Faseb J., 26, 2012
|
|
