3J7A
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, small subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
8A25
 
 | | Lysophospholipase PlaA from Legionella pneumophila str. Corby - complex with PEG fragment | | Descriptor: | (R,R)-2,3-BUTANEDIOL, IODIDE ION, Lysophospholipase A, ... | | Authors: | Diwo, M.G, Blankenfeldt, W. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-function relationships underpin disulfide loop cleavage-dependent activation of Legionella pneumophila lysophospholipase A PlaA.
Mol.Microbiol., 121, 2024
|
|
8A26
 
 | | Lysophospholipase PlaA from Legionella pneumophila str. Corby - complex with palmitate | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Lysophospholipase A, MALONIC ACID, ... | | Authors: | Diwo, M.G, Blankenfeldt, W. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function relationships underpin disulfide loop cleavage-dependent activation of Legionella pneumophila lysophospholipase A PlaA.
Mol.Microbiol., 121, 2024
|
|
2HB1
 
 | | Crystal Structure of PTP1B with Monocyclic Thiophene Inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K, Lee, J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3DY0
 
 | | Crystal Structure of Cleaved PCI Bound to Heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-terminus Plasma serine protease inhibitor, GLYCEROL, ... | | Authors: | Li, W, Huntington, J.A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The heparin binding site of protein C inhibitor is protease-dependent.
J.Biol.Chem., 283, 2008
|
|
3ZQI
 
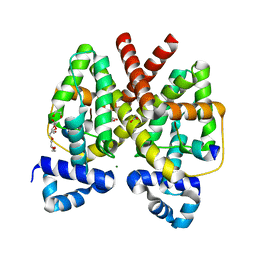 | | Structure of Tetracycline repressor in complex with inducer peptide- TIP2 | | Descriptor: | 1,2-ETHANEDIOL, INDUCER PEPTIDE TIP2, MAGNESIUM ION, ... | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
3K5N
 
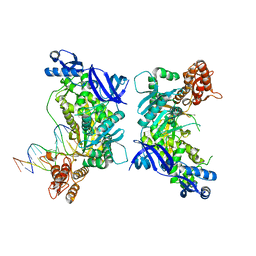 | | Crystal structure of E.coli Pol II-abasic DNA binary complex | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*TP*GP*(3DR)*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*G)-3'), DNA polymerase II | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
3DZQ
 
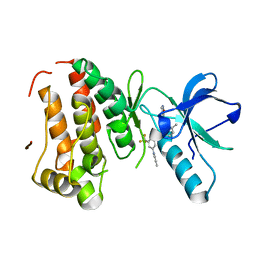 | | Human EphA3 kinase domain in complex with inhibitor AWL-II-38.3 | | Descriptor: | EPH receptor A3, N-[2-methyl-5-({[3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]carbonyl}amino)phenyl]isoxazole-5-carboxamide | | Authors: | Walker, J.R, Syeda, F, Gray, N, Mansoor, W, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinase Domain of Human Ephrin Type-A Receptor 3 (Epha3) in Complex with ALW-II-38-3.
To be Published
|
|
2HEH
 
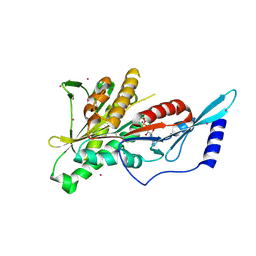 | | Crystal Structure of the KIF2C motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF2C protein, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the kif2c motor domain
to be published
|
|
3ZPX
 
 | |
1OPK
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
2GTT
 
 | | Crystal structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H. | | Deposit date: | 2006-04-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structure of the Rabies Virus Nucleoprotein-RNA Complex
Science, 313, 2006
|
|
3ZPH
 
 | |
5BWO
 
 | | Crystal Structure of Human SIRT3 in Complex with a Palmitoyl H3K9 Peptide | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, PALMITIC ACID, ... | | Authors: | Gai, W, Jiang, H, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
3J9Z
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
2HI9
 
 | |
3ZW8
 
 | | Crystal Structure Of Rat Peroxisomal Multifunctional Enzyme Type 1 (rpMFE1) In Apo Form | | Descriptor: | GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, SULFATE ION | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3JAV
 
 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
5BXY
 
 | | Crystal structure of RNA methyltransferase from Salinibacter ruber in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA methyltransferase, ... | | Authors: | Handing, K.B, LaRowe, C, Shabalin, I.G, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of RNA methylase family protein from Salinibacterruber in complex with S-Adenosyl-L-homocysteine.
to be published
|
|
3ZWQ
 
 | |
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
8A8E
 
 | | PPSA C terminal octahedral structure | | Descriptor: | Phosphoenolpyruvate synthase | | Authors: | Song, W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Survival strategies in the heat Lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermophilic PEP-synthase
To Be Published
|
|
2HGZ
 
 | | Crystal structure of a p-benzoyl-L-phenylalanyl-tRNA synthetase | | Descriptor: | PARA-(BENZOYL)-PHENYLALANINE, Tyrosyl-tRNA synthetase | | Authors: | Liu, W, Alfonta, L, Mack, A.V, Schultz, P.G. | | Deposit date: | 2006-06-27 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
1OH2
 
 | |
3DZO
 
 | | Crystal structure of a rhoptry kinase from toxoplasma gondii | | Descriptor: | MAGNESIUM ION, Rhoptry kinase domain | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Ni, S, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sibley, D, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
