1ETS
 
 | |
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LFF
 
 | | Solution structure of Diiron protein in presence of 8 eq Zn2+, Northeast Structural Genomics consortium target OR21 | | Descriptor: | Diiron protein, ZINC ION | | Authors: | Pires, M, Wu, Y, Mills, J.L, Reig, A, Englander, W, Degrado, W, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Diiron protein in presence of 8 eq Zn2+
To be Published
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
8P4N
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated disordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8P4P
 
 | | Structure average of GroEL14 complexes found in the cytosol of Escherichia coli overexpressing GroEL obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
7OZQ
 
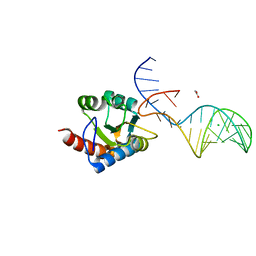 | | Crystal structure of archaeal L7Ae bound to eukaryotic kink-loop | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites.
Sci Rep, 11, 2021
|
|
8QXU
 
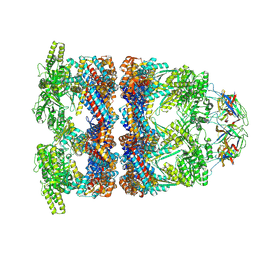 | | In situ structure average of GroEL14-GroES7 complexes with wide GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8QXT
 
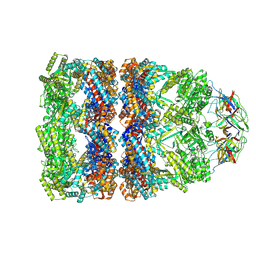 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with narrow GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8QXV
 
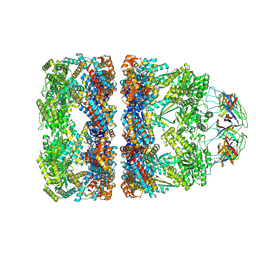 | | In situ structure average of GroEL14-GroES7 complexes with narrow GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
5DYK
 
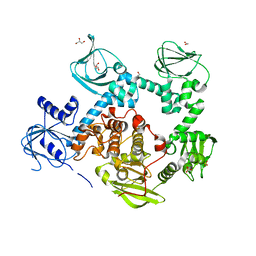 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium falciparum - Apo form | | Descriptor: | 1,2-ETHANEDIOL, CGMP-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PV2
 
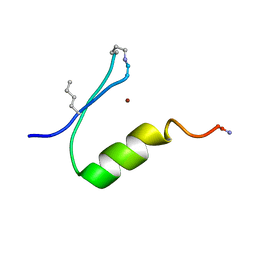 | |
5ZE0
 
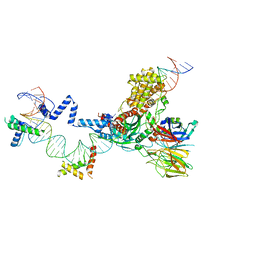 | | Hairpin Forming Complex, RAG1/2-Nicked(with Dideoxy) 12RSS/23RSS complex in Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (39-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
6PV0
 
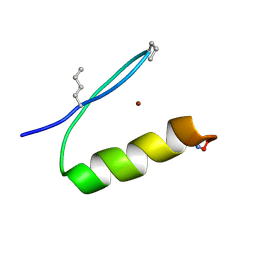 | |
7OT6
 
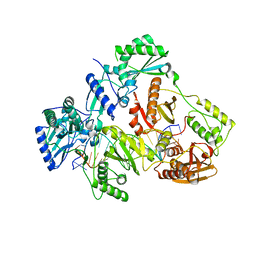 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND inhibitor RMC-282 | | Descriptor: | (R)-N-(1-(6-amino-9H-purin-9-yl)propan-2-yl)-N-(2-phosphonoethyl)glycine, DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTX
 
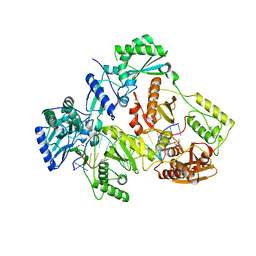 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-257 | | Descriptor: | (S)-2-((3-(6-amino-9H-purin-9-yl)propyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OUT
 
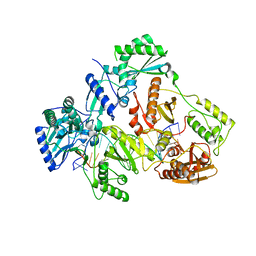 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-264 | | Descriptor: | (S)-2-(3-(6-amino-9H-purin-9-yl)propoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTK
 
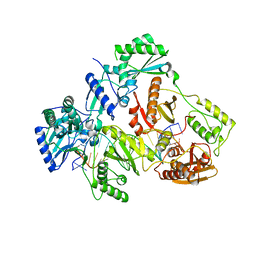 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-233 | | Descriptor: | (2~{R})-2-[2-(6-aminopurin-9-yl)ethylamino]-3-phosphono-propanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
8YVV
 
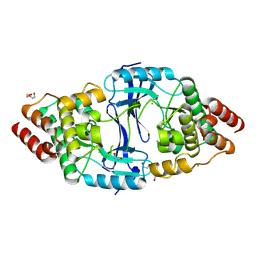 | | The Crystal Structure of BTK from Biortus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of BTK from Biortus.
To Be Published
|
|
7OTA
 
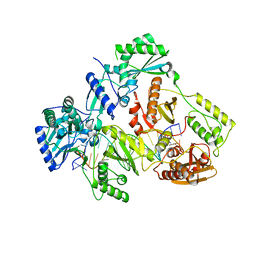 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-230 | | Descriptor: | ((2-(6-amino-9H-purin-9-yl)ethyl)-L-seryl)phosphoramidic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTN
 
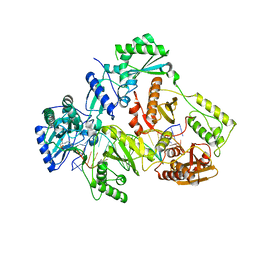 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | Descriptor: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTZ
 
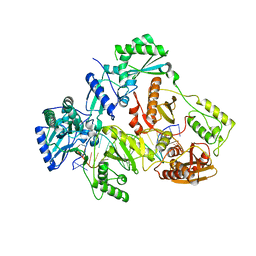 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-259 | | Descriptor: | (S)-2-(2-(6-amino-9H-purin-9-yl)ethoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
