1BS9
 
 | | ACETYLXYLAN ESTERASE FROM P. PURPUROGENUM REFINED AT 1.10 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3JTK
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055 | | Descriptor: | (2R)-3-benzyl-2-(2-bromo-4-hydroxy-5-methoxyphenyl)-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055
To be Published
|
|
3K58
 
 | | Crystal structure of E.coli Pol II-normal DNA-dTTP ternary complex | | Descriptor: | DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*AP*AP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), DNA polymerase II, ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II
Cell(Cambridge,Mass.), 139, 2009
|
|
3K5L
 
 | | Crystal structure of E.coli Pol II-abasic DNA-dATP Lt(0, 3) ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*AP*TP*(3DR)P*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
1Y3N
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
2Q3F
 
 | | X-ray crystal structure of putative human Ras-related GTP binding D in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related GTP-binding protein D | | Authors: | Mulichak, A.M, Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Keefe, L.J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Ras-related GTP-binding D.
To be published
|
|
1Y74
 
 | | Solution Structure of mLin-2/mLin-7 L27 Domain Complex | | Descriptor: | Peripheral plasma membrane protein CASK, lin 7 homolog b | | Authors: | Feng, W, Long, J.-F, Zhang, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A unified assembly mode revealed by the structures of tetrameric L27 domain complexes formed by mLin-2/mLin-7 and Patj/Pals1 scaffold proteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y6H
 
 | | Crystal structure of LIPDF | | Descriptor: | FORMIC ACID, GLYCINE, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-12-06 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique structural characteristics of peptide deformylase from pathogenic bacterium Leptospira interrogans
J.Mol.Biol., 339, 2004
|
|
3KTT
 
 | | Atomic model of bovine TRiC CCT2(beta) subunit derived from a 4.0 Angstrom cryo-EM map | | Descriptor: | T-complex protein 1 subunit beta | | Authors: | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3K19
 
 | | OmpF porin | | Descriptor: | Outer membrane protein F | | Authors: | Kefala, G, Ahn, C, Krupa, M, Maslennikov, I, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-09-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structures of the OmpF porin crystallized in the presence of foscholine-12.
Protein Sci., 19, 2010
|
|
3JAE
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
3K9O
 
 | | The crystal structure of E2-25K and UBB+1 complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
2WJJ
 
 | | Structure and function of the FeoB G-domain from Methanococcus jannaschii | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B HOMOLOG, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Koester, S, Wehner, M, Herrmann, C, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2009-05-26 | | Release date: | 2009-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure and Function of the Feob G-Domain from Methanococcus Jannaschii
J.Mol.Biol., 392, 2009
|
|
2WL8
 
 | | X-ray crystal structure of Pex19p | | Descriptor: | PEROXISOMAL BIOGENESIS FACTOR 19 | | Authors: | Schueller, N, Holton, S.J, Stanley, W.A, Song, Y.H, Konarev, P, Roessle, M, Erdmann, R, Schliebs, W, Wilmanns, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Peroxisomal Receptor Pex19P Forms a Helical Mpts Recognition Domain.
Embo J., 29, 2010
|
|
2X6L
 
 | |
1CFQ
 
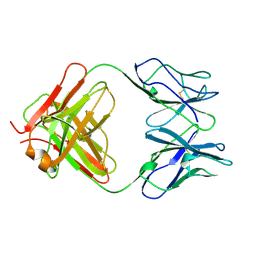 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 | | Descriptor: | PROTEIN (IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN)), PROTEIN (IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN)) | | Authors: | Keitel, T, Kramer, A, Wessner, H, Scholz, C, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 1999-03-19 | | Release date: | 1999-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity.
Cell(Cambridge,Mass.), 91, 1997
|
|
2X3V
 
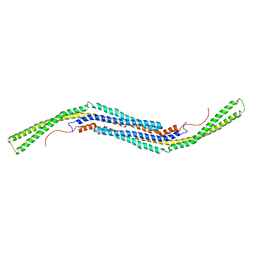 | | Structure of The F-BAR Domain of Mouse Syndapin I | | Descriptor: | PROTEIN KINASE C AND CASEIN KINASE SUBSTRATE IN NEURONS PROTEIN 1 | | Authors: | Ma, Q, Rao, Y, Vahedi-Faridi, A, Saenger, W, Haucke, V. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular Basis for SH3 Domain Regulation of F-Bar-Mediated Membrane Deformation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1ZB7
 
 | | Crystal Structure of Botulinum Neurotoxin Type G Light Chain | | Descriptor: | CITRATE ANION, ZINC ION, neurotoxin | | Authors: | Arndt, J.W, Yu, W, Bi, F, Stevens, R.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of botulinum neurotoxin type g light chain: serotype divergence in substrate recognition
Biochemistry, 44, 2005
|
|
2X69
 
 | |
1Z28
 
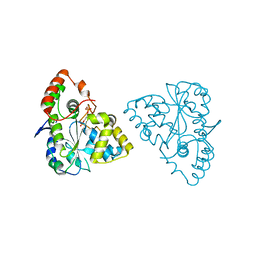 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phenol-sulfating phenol sulfotransferase 1 | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1ZOD
 
 | |
1ZN1
 
 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1Y79
 
 | | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp in Complex with a Peptidic Inhibitor | | Descriptor: | ASPARTIC ACID, GLYCINE, LYSINE, ... | | Authors: | Comellas-Bigler, M, Lang, R, Bode, W, Maskos, K. | | Deposit date: | 2004-12-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp: Further Indication of a Ligand-dependant Hinge Movement Mechanism
J.Mol.Biol., 349, 2005
|
|
2RGH
 
 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-01 | | Release date: | 2008-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
