4BV5
 
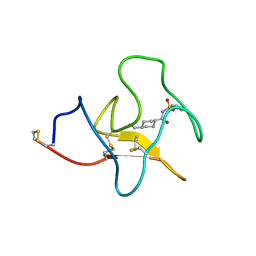 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
1PUA
 
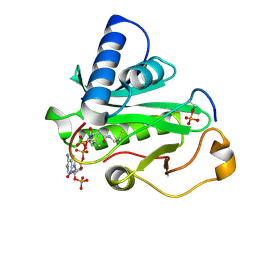 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a Phosphorylated, 19-residue Histone H3 peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
8XBI
 
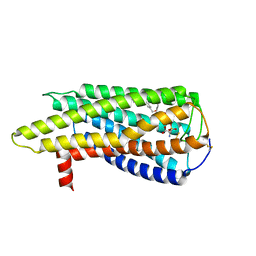 | | Human GPR34 -Gi complex bound to M1, receptor focused | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XFM
 
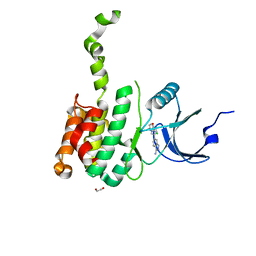 | | The Crystal Structure of MNK2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-azanyl-1~{H}-indazol-6-yl)-1-[(3-chlorophenyl)methyl]pyridin-2-one, MAP kinase-interacting serine/threonine-protein kinase 2, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-12-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of MNK2 from Biortus.
To Be Published
|
|
3T4Z
 
 | | Crystal Structure of NaK2K Channel Y55W Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4HXA
 
 | | Crystal structure of 13D9 FAB | | Descriptor: | 13D9 FAB HEAVY CHAIN, 13D9 FAB LIGHT CHAIN | | Authors: | Johal, A.R, Jarrell, H.C, Khieu, N.H, Letts, J.A, Landry, R.C, Jachymek, W, Yang, Q, Jennings, H.J, Brisson, J.R, Evans, S.V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The antigen-binding site of an N-propionylated polysialic acid-specific antibody protective against group B meningococci is consistent with extended epitopes.
Glycobiology, 23, 2013
|
|
4CCZ
 
 | | Crystal structure of human 5-methyltetrahydrofolate-homocysteine methyltransferase, the homocysteine and folate binding domains | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, METHIONINE SYNTHASE | | Authors: | Vollmar, M, Kiyani, W, Krojer, T, Goubin, S, Burgess-Brown, N, von Delft, F, Oppermann, U, Edwards, A, Arrowsmith, C, Bountra, C, Yue, W.W. | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human 5-Methyltetrahydrofolate-Homocysteine Methyltransferase, the Homocysteine and Folate Binding Domains
To be Published
|
|
4HZH
 
 | | Structure of recombinant Gla-domainless prothrombin mutant S525A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Niu, W, Gohara, D.W, Chen, Z, Di Cera, E. | | Deposit date: | 2012-11-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of prothrombin reveals conformational flexibility and mechanism of activation.
J.Biol.Chem., 288, 2013
|
|
4I11
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates. | | Descriptor: | Beta-secretase 1, N-(3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-phenylalanine, ZINC ION | | Authors: | Bowers, B, Xu, Y, Yuan, S, Probst, G.D, Hom, R.K, Chan, W, Konradi, A.W, Sham, H.L, Zhu, Y.L, Beroza, P, Pan, H, Brecht, E, Yao, N, Lougheed, J, Artis, D.R, Tam, D, Bova, M. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4BVV
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4MRY
 
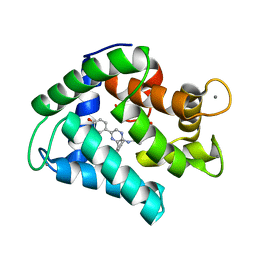 | | Crystal Structure of Ca(2+)- discharged Y138F obelin mutant from Obelia longissima at 1.30 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4I6J
 
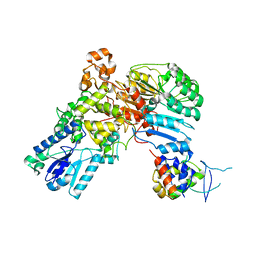 | | A ubiquitin ligase-substrate complex | | Descriptor: | Cryptochrome-2, F-box/LRR-repeat protein 3, S-phase kinase-associated protein 1 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I76
 
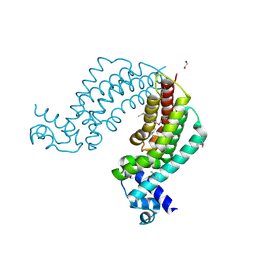 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
4CAV
 
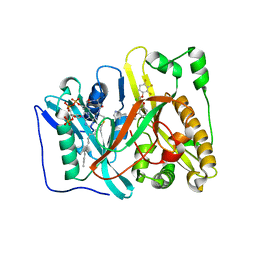 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
3TFZ
 
 | | Crystal structure of Zhui aromatase/cyclase from Streptomcyes sp. R1128 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclase, POTASSIUM ION | | Authors: | Ames, B.D, Lee, M.Y, Moody, C, Zhang, W, Tang, Y, Wong, S.K, Tsai, S.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Characterization of ZhuI Aromatase/Cyclase from the R1128 Polyketide Pathway.
Biochemistry, 50, 2011
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4CCU
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy) pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol | | Descriptor: | 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4N9Y
 
 | |
4NA0
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
8TW0
 
 | | Crystal Structure of a synthetic ABC heterotrimeric Collagen-like Peptide at 1.53 A | | Descriptor: | Collagen Mimetic Peptide A, Collagen Mimetic Peptide B, Collagen Mimetic Peptide C, ... | | Authors: | Miller, M.D, Cole, C.C, Xu, W, Walker, D.R, Hulgan, S.A.H, Pogostin, B.H, Swain, J.W.R, Duella, R, Misiura, M, Wang, X, Kolomeisky, A.B, Phillips Jr, G.N, Hartgerink, J.D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Heterotrimeric Collagen Helix with High Specificity of Assembly Results in a Rapid Rate of Folding
Nat.Chem., 2024
|
|
3SPW
 
 | | Structure of Osh4p/Kes1p in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Protein KES1 | | Authors: | Delfosse, V, de Saint-Jean, M, Douguet, D, Antonny, B, Drin, G, Bourguet, W. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers.
J.Cell Biol., 195, 2011
|
|
4IAQ
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IB4
 
 | | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, C, Katritch, V, Han, G.W, Huang, X, Vardy, E, McCorvy, J.D, Jiang, Y, Chu, M, Siu, F.Y, Liu, W, Xu, H.E, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural features for functional selectivity at serotonin receptors.
Science, 340, 2013
|
|
3SSP
 
 | |
4BVW
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
