3TYK
 
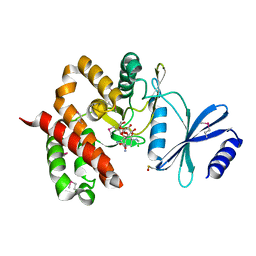 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | 分子名称: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | 著者 | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-26 | | 公開日 | 2011-10-12 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | 分子名称: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | 著者 | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
3U9I
 
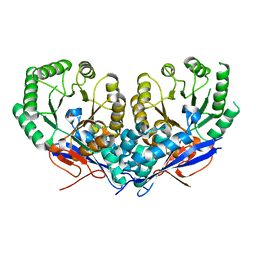 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Roseiflexus sp. | | 分子名称: | Mandelate racemase/muconate lactonizing enzyme, C-terminal domain protein, SULFATE ION | | 著者 | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-10-19 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Roseiflexus sp.
TO BE PUBLISHED
|
|
3TOX
 
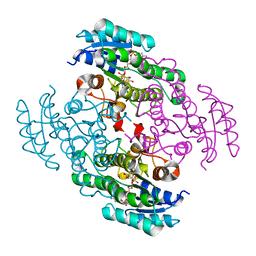 | | Crystal structure of a short chain dehydrogenase in complex with NAD(P) from Sinorhizobium meliloti 1021 | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-09-06 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure of a short chain dehydrogenase in complex with NAD(P) from Sinorhizobium meliloti 1021
To be Published
|
|
3TT0
 
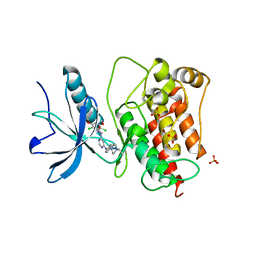 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | 分子名称: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | 著者 | Bussiere, D.E, Murray, J.M, Shu, W. | | 登録日 | 2011-09-13 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
3TXV
 
 | | Crystal structure of a probable tagatose 6 phosphate kinase from Sinorhizobium meliloti 1021 | | 分子名称: | Probable tagatose 6-phosphate kinase | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-09-23 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of a probable tagatose 6 phosphate kinase from Sinorhizobium meliloti 1021
To be Published
|
|
3U3X
 
 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 | | 分子名称: | ACETATE ION, Oxidoreductase | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-10-06 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021
To be Published
|
|
3U4Q
 
 | | Structure of AddAB-DNA complex at 2.8 angstroms | | 分子名称: | 1,2-ETHANEDIOL, ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, ... | | 著者 | Saikrishnan, K, Krajewski, W, Wigley, D. | | 登録日 | 2011-10-10 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
7THS
 
 | | Macrocyclic plasmin inhibitor | | 分子名称: | (2S)-butane-1,2-diol, (6S,9R,20R,23S)-N-{[4-(aminomethyl)phenyl]methyl}-20-[(benzenesulfonyl)amino]-3,13,21-trioxo-2,6,9,14,22-pentaazatetracyclo[23.2.2.2~6,9~.2~15,18~]tritriaconta-1(27),15,17,25,28,30-hexaene-23-carboxamide, Plasminogen, ... | | 著者 | Guojie, W. | | 登録日 | 2022-01-12 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7UAH
 
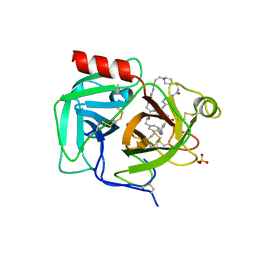 | | Macrocyclic plasmin inhibitor | | 分子名称: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | 著者 | Guojie, W. | | 登録日 | 2022-03-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7VRT
 
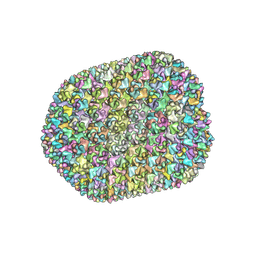 | | The unexpanded head structure of phage T4 | | 分子名称: | Capsid vertex protein, Major capsid protein | | 著者 | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | 登録日 | 2021-10-24 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VS5
 
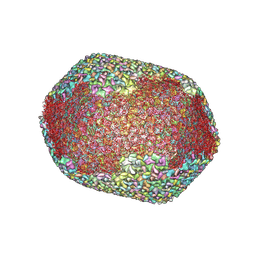 | | The expanded head structure of phage T4 | | 分子名称: | Capsid vertex protein, Major capsid protein, Small outer capsid protein | | 著者 | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | 登録日 | 2021-10-25 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WIG
 
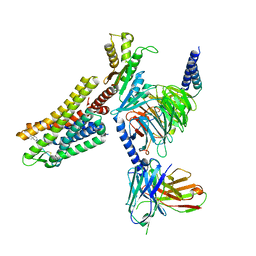 | | Cryo-EM structure of the L-054,264-bound human SSTR2-Gi1 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Chen, L, Wang, W, Dong, Y, Shen, D, Guo, J, Qin, J, Zhang, H, Shen, Q, Zhang, Y, Mao, C. | | 登録日 | 2022-01-03 | | 公開日 | 2022-06-01 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of the endogenous peptide- and selective non-peptide agonist-bound SSTR2 signaling complexes.
Cell Res., 32, 2022
|
|
7WIC
 
 | | Cryo-EM structure of the SS-14-bound human SSTR2-Gi1 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Chen, L, Wang, W, Dong, Y, Shen, D, Guo, J, Qin, J, Zhang, H, Shen, Q, Zhang, Y, Mao, C. | | 登録日 | 2022-01-03 | | 公開日 | 2022-06-01 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of the endogenous peptide- and selective non-peptide agonist-bound SSTR2 signaling complexes.
Cell Res., 32, 2022
|
|
7VT0
 
 | | Dimer structure of SORLA | | 分子名称: | Sortilin-related receptor | | 著者 | Xi, Z, Cang, W, Chuang, L. | | 登録日 | 2021-10-27 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-05-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
7UYK
 
 | | Structure of RNF31 in complex with FP06655, a Helicon Polypeptide | | 分子名称: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06655, ... | | 著者 | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | 登録日 | 2022-05-06 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UY2
 
 | | Structure of RNF31 in complex with FP06649, a Helicon Polypeptide | | 分子名称: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06649, ... | | 著者 | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | 登録日 | 2022-05-06 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UYJ
 
 | | Structure of RNF31 in complex with FP06652, a Helicon Polypeptide | | 分子名称: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06652, ... | | 著者 | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | 登録日 | 2022-05-06 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VMU
 
 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | 分子名称: | Spike protein S1, scFv E4 | | 著者 | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | 登録日 | 2021-10-09 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | 著者 | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | 登録日 | 2022-03-17 | | 公開日 | 2023-03-22 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
7XLB
 
 | |
7C97
 
 | |
7CHW
 
 | |
7C7D
 
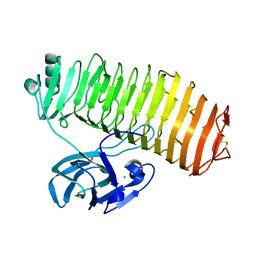 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | 分子名称: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | 著者 | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | 登録日 | 2020-05-25 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7D5T
 
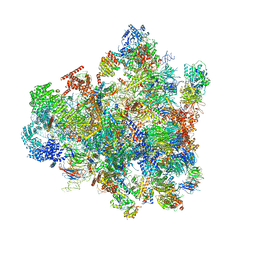 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
