4N92
 
 | |
8XJ6
 
 | | The Cryo-EM structure of MPXV E5 apo conformation | | 分子名称: | AMP PHOSPHORAMIDATE, Monkeypox virus E5, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Zhang, W, Liu, Y, Gao, H, Gan, J. | | 登録日 | 2023-12-20 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
4N3C
 
 | |
4IX1
 
 | | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205 | | 分子名称: | PHOSPHATE ION, hypothetical protein | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-01-24 | | 公開日 | 2013-02-06 | | 最終更新日 | 2013-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205
To be Published
|
|
4ITH
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-1 analog | | 分子名称: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1, ... | | 著者 | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | 登録日 | 2013-01-18 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
8XIG
 
 | |
4N9C
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8XJ7
 
 | | The Cryo-EM structure of MPXV E5 in complex with DNA | | 分子名称: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | 著者 | Zhang, W, Liu, Y, Gao, H, Gan, J. | | 登録日 | 2023-12-20 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
4NA6
 
 | |
4E59
 
 | |
3QCA
 
 | |
4E5T
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200750) from Labrenzia alexandrii DFL-11 | | 分子名称: | MAGNESIUM ION, Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | 著者 | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-03-14 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a putative MR/ML enzyme from Labrenzia alexandrii DFL-11
to be published
|
|
8YC0
 
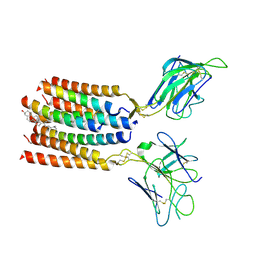 | | T cell receptor V delta2 V gamma9 in GDN | | 分子名称: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | 著者 | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | 登録日 | 2024-02-17 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
5UXI
 
 | |
4LUQ
 
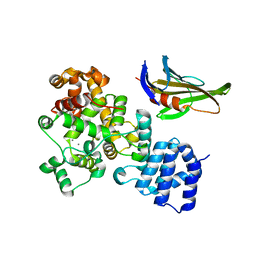 | |
4ECT
 
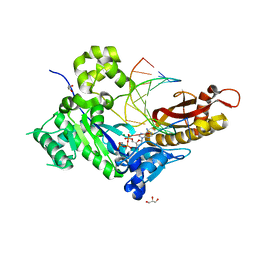 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 140 sec | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | 著者 | Nakamura, T, Zhao, Y, Yang, W. | | 登録日 | 2012-03-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.795 Å) | | 主引用文献 | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED0
 
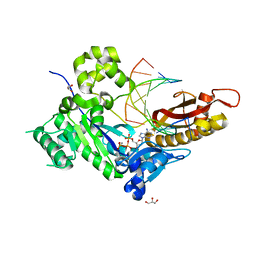 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 6.8 (Na+ MES) with 1 Ca2+ ion | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | 著者 | Nakamura, T, Zhao, Y, Yang, W. | | 登録日 | 2012-03-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4LW5
 
 | | Crystal structure of all-trans green fluorescent protein | | 分子名称: | Green fluorescent protein | | 著者 | Rosenman, D.J, Huang, Y.-M, Xia, K, Vanroey, P, Colon, W, Bystroff, C. | | 登録日 | 2013-07-26 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Green-lighting green fluorescent protein: Faster and more efficient folding by eliminating a cis-trans peptide isomerization event.
Protein Sci., 23, 2014
|
|
4LWW
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, N-(4-(phenylsulfonyl)benzyl)-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | 登録日 | 2013-07-28 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4J1G
 
 | | Leanyer orthobunyavirus nucleoprotein-ssRNA complex | | 分子名称: | Nucleocapsid, RNA (45-MER) | | 著者 | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | 登録日 | 2013-02-01 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.789 Å) | | 主引用文献 | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DCL
 
 | | Computationally Designed Self-assembling tetrahedron protein, T308, Crystallized in space group F23 | | 分子名称: | Putative acetyltransferase SACOL2570 | | 著者 | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2012-01-17 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4M0I
 
 | |
4DUN
 
 | | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630) | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, Putative phenazine biosynthesis PhzC/PhzF protein, ... | | 著者 | Brunzelle, J.S, Wawrzak, W, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-02-22 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630)
To be Published
|
|
