7GG7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with CHO-MSK-00c5269a-2 (Mpro-x12177) | | 分子名称: | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-(4-methoxyphenyl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGM
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-afd4d4fd-2 (Mpro-x12677) | | 分子名称: | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.839 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGO
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-8b8a49e1-4 (Mpro-x12682) | | 分子名称: | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.687 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4HCB
 
 | | The metal-free form of crystal structure of E.coli ExoI-ssDNA complex | | 分子名称: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, GLYCEROL, ... | | 著者 | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | 登録日 | 2012-09-29 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
4HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, 25 STRUCTURES | | 分子名称: | HEMATOPOIETIC CELL KINASE | | 著者 | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | 登録日 | 1998-03-09 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
4GR1
 
 | |
4HMS
 
 | | Crystal structure of PhzG from Pseudomonas fluorescens 2-79 in complex with a second FMN in the substrate binding site | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | 著者 | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMD
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | 分子名称: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | 著者 | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HP3
 
 | | Crystal structure of Tet3 in complex with a CpG dsDNA | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), LOC100036628 protein, UNKNOWN ATOM OR ION, ... | | 著者 | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2012-10-23 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | 分子名称: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | 著者 | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | 登録日 | 2012-10-15 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
4HIC
 
 | |
1DG0
 
 | | NMR STRUCTURE OF DES[GLY1]-CONTRYPHAN-R CYCLIC PEPTIDE (MAJOR FORM) | | 分子名称: | DES[GLY1]-CONTRYPHAN-R | | 著者 | Pallaghy, P.K, He, W, Jimenez, E.C, Olivera, B.M, Norton, R.S. | | 登録日 | 1999-11-22 | | 公開日 | 2003-09-09 | | 最終更新日 | 2020-06-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the contryphan family of cyclic peptides. Role of electrostatic interactions in cis-trans isomerism
Biochemistry, 39, 2000
|
|
4HME
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | 著者 | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | 登録日 | 2012-10-18 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1DCH
 
 | | CRYSTAL STRUCTURE OF DCOH, A BIFUNCTIONAL, PROTEIN-BINDING TRANSCRIPTION COACTIVATOR | | 分子名称: | DCOH (DIMERIZATION COFACTOR OF HNF-1), SULFATE ION | | 著者 | Endrizzi, J.A, Cronk, J.D, Wang, W, Crabtree, G.R, Alber, T. | | 登録日 | 1995-01-24 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science, 268, 1995
|
|
1DFF
 
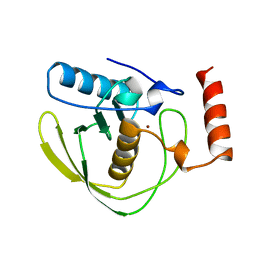 | | PEPTIDE DEFORMYLASE | | 分子名称: | PEPTIDE DEFORMYLASE, ZINC ION | | 著者 | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | 登録日 | 1997-08-19 | | 公開日 | 1998-09-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
7ZEI
 
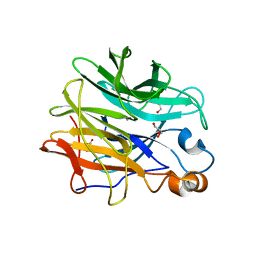 | | Thermostable GH159 glycoside hydrolase from Caldicellulosiruptor at 1.7 A | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | 著者 | Baudrexl, M, Fida, T, Berk, B, Schwarz, W, Zverlov, V.V, Groll, M, Liebl, W. | | 登録日 | 2022-03-31 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Biochemical and Structural Characterization of Thermostable GH159 Glycoside Hydrolases Exhibiting alpha-L-Arabinofuranosidase Activity.
Front Mol Biosci, 9, 2022
|
|
1DP3
 
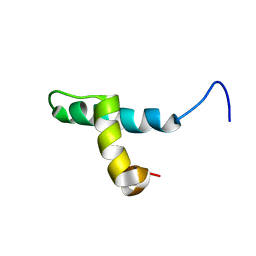 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF THE TRAM PROTEIN | | 分子名称: | TRAM PROTEIN | | 著者 | Stockner, T, Plugariu, C, Koraimann, G, Hoegenauer, G, Bermel, W, Prytulla, S, Sterk, H. | | 登録日 | 1999-12-23 | | 公開日 | 2001-04-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA-binding domain of TraM.
Biochemistry, 40, 2001
|
|
3NIL
 
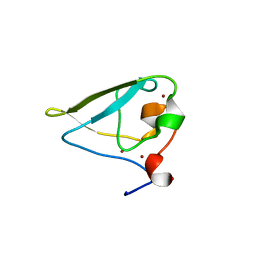 | | The structure of UBR box (RDAA) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
 | | The structure of UBR box (REAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1FP7
 
 | | MONOVALENT CATION BINDING SITES IN N10-FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | 分子名称: | FORMATE--TETRAHYDROFOLATE LIGASE, POTASSIUM ION, SULFATE ION | | 著者 | Radfar, R, Leaphart, A, Brewer, J.M, Minor, W, Odom, J.D. | | 登録日 | 2000-08-30 | | 公開日 | 2001-08-30 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Cation binding and thermostability of FTHFS monovalent cation binding sites and thermostability of N10-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
4GVL
 
 | | Crystal Structure of the GsuK RCK domain | | 分子名称: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, TrkA domain protein, ... | | 著者 | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
1FI9
 
 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | 分子名称: | CYTOCHROME C, HEME C, IMIDAZOLE | | 著者 | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | 登録日 | 2000-08-03 | | 公開日 | 2000-08-23 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
4H33
 
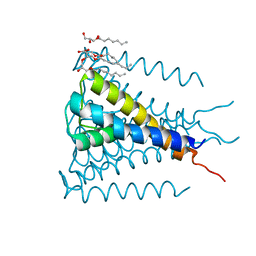 | | Crystal structure of a voltage-gated K+ channel pore module in a closed state in lipid membranes, tetragonal crystal form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lmo2059 protein, POTASSIUM ION | | 著者 | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
1FO7
 
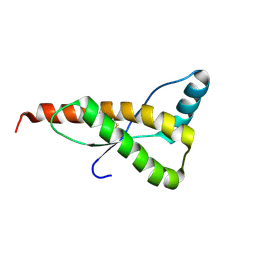 | | HUMAN PRION PROTEIN MUTANT E200K FRAGMENT 90-231 | | 分子名称: | PRION PROTEIN | | 著者 | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | 登録日 | 2000-08-25 | | 公開日 | 2000-09-21 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
