7YNI
 
 | | Structure of human SGLT1-MAP17 complex bound with substrate 4D4FDG in the occluded conformation | | Descriptor: | (2R,3R,4R,5S,6R)-5-fluoranyl-6-(hydroxymethyl)oxane-2,3,4-triol, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-07-31 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of human SGLT in the occluded state reveal conformational changes during sugar transport.
Nat Commun, 14, 2023
|
|
7YL5
 
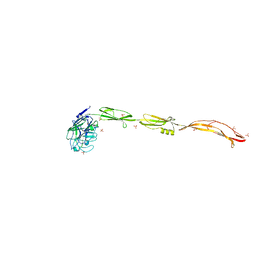 | | Cell surface protein YwfG protein complexed with mannose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
7YL6
 
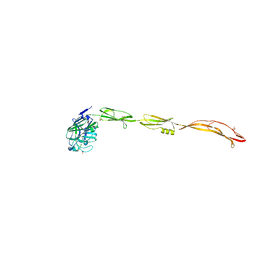 | | Cell surface protein YwfG protein complexed with alpha-1,2-mannobiose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
7YL4
 
 | | Cell surface protein YwfG protein (apo form) | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
4IE7
 
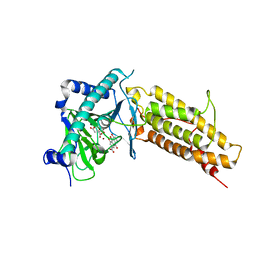 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with citrate and rhein (RHN) | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, CITRATE ANION, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6004 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
7YKO
 
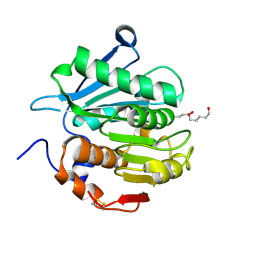 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
7YKQ
 
 | |
7YKP
 
 | | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol | | Descriptor: | GLYCEROL, Triacylglycerol lipase | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol
To Be Published
|
|
4IE6
 
 | |
7YLQ
 
 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
4IDZ
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with N-oxalylglycine (NOG) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
7YRS
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YW1
 
 | | crystal structure of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | crystal structure of UBE2O
To Be Published
|
|
4LFM
 
 | |
4LFN
 
 | |
4KGS
 
 | | Backbone Modifications in the Protein GB1 Loops: beta-3-Val21, beta-3-Asp40 | | Descriptor: | GLYCEROL, Streptococcal Protein GB1 Backbone Modified Variant: beta-3-Val21, beta-3-Asp40 | | Authors: | Reinert, Z.E, Lengyel, G.A, Horne, W.S. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protein-like Tertiary Folding Behavior from Heterogeneous Backbones.
J.Am.Chem.Soc., 135, 2013
|
|
4KCA
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4KGR
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Lys31, beta-3-Asn35 | | Descriptor: | GLYCEROL, Streptococcal Protein GB1 Backbone Modified Variant: beta-3-Ala24, beta-3-Lys28, ... | | Authors: | Reinert, Z.E, Lengyel, G.A, Horne, W.S. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-like Tertiary Folding Behavior from Heterogeneous Backbones.
J.Am.Chem.Soc., 135, 2013
|
|
4LFK
 
 | |
4LFL
 
 | |
6WZX
 
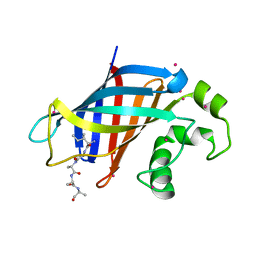 | | GID4 in complex with IGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
6WIB
 
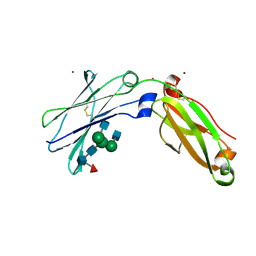 | | Next generation monomeric IgG4 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 4, ZINC ION | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W, van Dyk, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6XL3
 
 | | Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | CHLORIDE ION, DECANE, Mastigocladopsis repens rhodopsin chloride pump, ... | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
6XRE
 
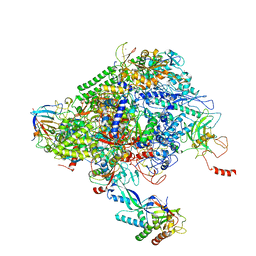 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
