3IYG
 
 | | Ca model of bovine TRiC/CCT derived from a 4.0 Angstrom cryo-EM map | | Descriptor: | T-complex protein 1 subunit, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-11-28 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2JPB
 
 | |
3IZM
 
 | | Mm-cpn wildtype with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
1Q55
 
 | | W-shaped trans interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
3J0C
 
 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3IZL
 
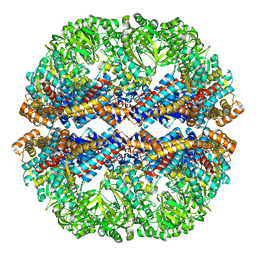 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3WMJ
 
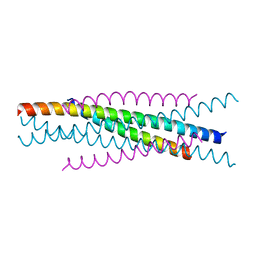 | | Crystal structure of EIAV vaccine gp45 | | Descriptor: | EIAV vaccine gp45 | | Authors: | Liu, X, Du, J, Qiao, W. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | A mutation associated with EIAV vaccine strain within heptad repeat of EIAV gp45 provides insight into vaccine development for HIV
To be Published
|
|
3IZN
 
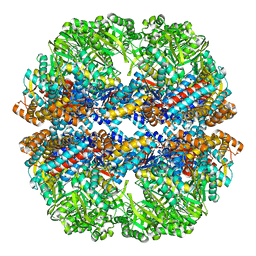 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3J0F
 
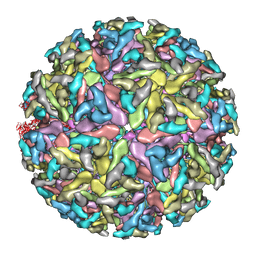 | | Sindbis virion | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Tang, J, Jose, J, Zhang, W, Chipman, P, Kuhn, R.J, Baker, T.S. | | Deposit date: | 2011-07-08 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus.
J.Mol.Biol., 414, 2011
|
|
4ZMK
 
 | |
3IZP
 
 | |
3CNI
 
 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
8A6U
 
 | | PcIDS1 in complex with Mg2+ | | Descriptor: | Isoprenyl diphosphate synthase, MAGNESIUM ION | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
2JFT
 
 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with sulfate | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SER-THR PHOSPHATASE MSPP, ... | | Authors: | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
2JFR
 
 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with phosphate at 0.83 A resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
3CON
 
 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
3COS
 
 | | Crystal structure of human class II alcohol dehydrogenase (ADH4) in complex with NAD and Zn | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alcohol dehydrogenase 4, ... | | Authors: | Kavanagh, K.L, Shafqat, N, Yue, W, von Delft, F, Bishop, S, Roos, A, Murray, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human class II alcohol dehydrogenase (ADH4) in complex with NAD and Zn.
To be Published
|
|
8A7K
 
 | | PcIDS1 in complex with Mg2+/Mn2+, IPP, and ZOL | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A6V
 
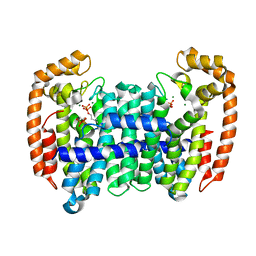 | | PcIDS1 in complex with Mg2+ and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Isoprenyl diphosphate synthase, MAGNESIUM ION | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
7ZPN
 
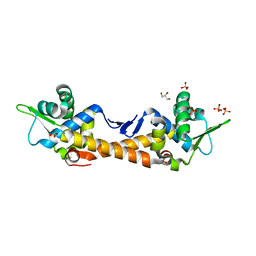 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
4Z3S
 
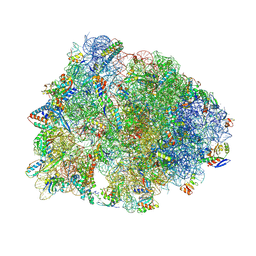 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
3J7L
 
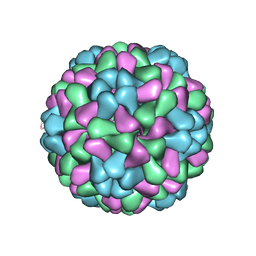 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
