7XUV
 
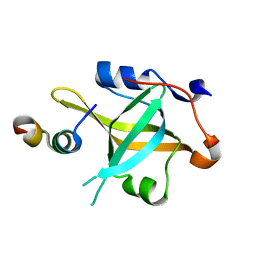 | | Crystal structure of RPA70N-RMI1 fusion | | Descriptor: | RecQ-mediated genome instability protein 1, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
7XUW
 
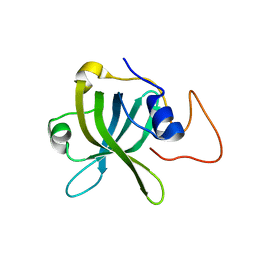 | | Crystal structure of RPA70N-BLMp2 fusion | | Descriptor: | fusion protein of Replication protein A 70 kDa DNA-binding subunit and Bloom syndrome protein | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
4DFF
 
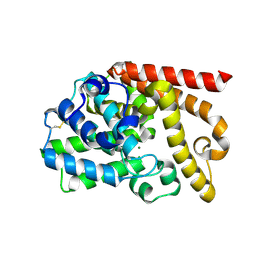 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | Descriptor: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6KE4
 
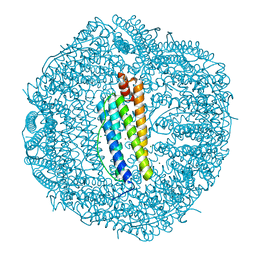 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
6KE2
 
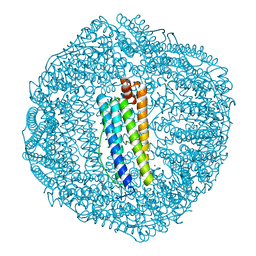 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
6LIX
 
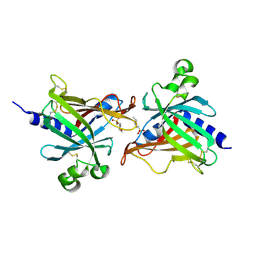 | | CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
6LIY
 
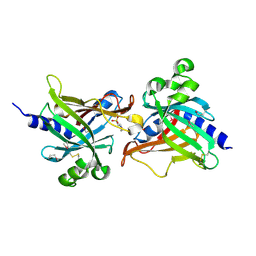 | | SeMet CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
6LYW
 
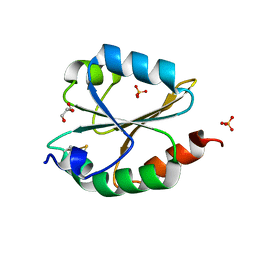 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
8W6U
 
 | | Ferritin from Ureaplasma diversum soaking in Fe2+ solution for 5 min | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8XB3
 
 | |
8XB4
 
 | |
8XWB
 
 | | Crystal structure of dinitrosyl iron units binding with human heavy chain Ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Gong, W.J, Wang, W.M, Wang, H.F. | | Deposit date: | 2024-01-16 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Insight into the photodynamic mechanism and protein binding of a nitrosyl iron-sulfur [Fe 2 S 2 (NO) 4 ] 2- cluster.
Spectrochim Acta A Mol Biomol Spectrosc, 320, 2024
|
|
7DIE
 
 | | Crystal structure of M. penetrans Ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, w.m, Zhang, y, Wang, h.f. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ferritin with Atypical Ferroxidase Centers Takes B-Channels as the Pathway for Fe 2+ Uptake from Mycoplasma .
Inorg.Chem., 60, 2021
|
|
8XB2
 
 | | Structure of radafaxine-bound state of the human Norepinephrine Transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GFP-MBP-solute carrier family 6 member 2,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter, Radafaxine | | Authors: | Wu, J.X, Ji, W.M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Substrate binding and inhibition mechanism of norepinephrine transporter.
Nature, 633, 2024
|
|
8ZTP
 
 | | Crystal structure of cysteine desulfurase Sufs from Mycoplasma Pneumonia | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, cysteine desulfurase | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
8ZTQ
 
 | | Crystal structure of Sufu from Mycoplasma Pneumonia | | Descriptor: | Nitrogen fixation protein NifU, ZINC ION | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
3EO3
 
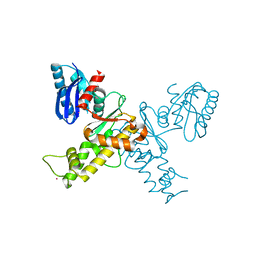 | | Crystal structure of the N-acetylmannosamine kinase domain of human GNE protein | | Descriptor: | Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Nedyalkova, L, Tong, Y, Rabeh, W.M, Hong, B, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the N-acetylmannosamine kinase domain of GNE.
Plos One, 4, 2009
|
|
3FEG
 
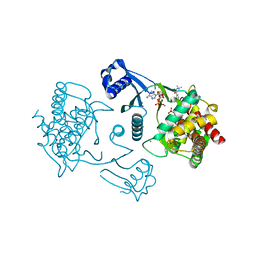 | | Crystal structure of human choline kinase beta in complex with phosphorylated hemicholinium-3 and adenosine nucleotide | | Descriptor: | (2S)-2-[4'-({dimethyl[2-(phosphonooxy)ethyl]ammonio}acetyl)biphenyl-4-yl]-2-hydroxy-4,4-dimethylmorpholin-4-ium, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B.S, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity.
J.Biol.Chem., 285, 2010
|
|
3G15
 
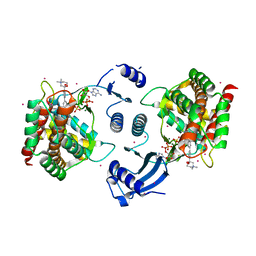 | | Crystal structure of human choline kinase alpha in complex with hemicholinium-3 and ADP | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), ADENOSINE-5'-DIPHOSPHATE, Choline kinase alpha, ... | | Authors: | Hong, B.S, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity.
J.Biol.Chem., 285, 2010
|
|
4KK1
 
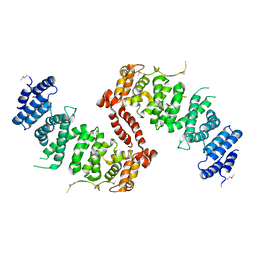 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
1T32
 
 | | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation | | Descriptor: | 2-[3-({METHYL[1-(2-NAPHTHOYL)PIPERIDIN-4-YL]AMINO}CARBONYL)-2-NAPHTHYL]-1-(1-NAPHTHYL)-2-OXOETHYLPHOSPHONIC ACID, Cathepsin G, SULFATE ION | | Authors: | de Garavilla, L, Greco, M.N, Giardino, E.C, Wells, G.I, Haertlein, B.J, Kauffman, J.A, Corcoran, T.W, Derian, C.K, Eckardt, A.J, Abraham, W.M, Sukumar, N, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E, Andrade-Gordon, P, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2004-04-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
J.Biol.Chem., 280, 2005
|
|
8H7V
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
