8PDI
 
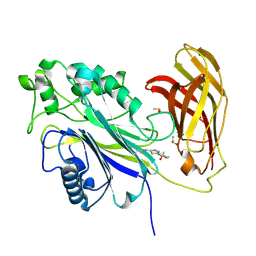 | | The phosphatase and C2 domains of SHIP1 with covalent Z1763271112 | | Descriptor: | (5-phenyl-1,3,4-thiadiazol-2-yl)methanimine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
1DLP
 
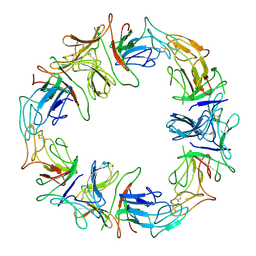 | | STRUCTURAL CHARACTERIZATION OF THE NATIVE FETUIN-BINDING PROTEIN SCILLA CAMPANULATA AGGLUTININ (SCAFET): A NOVEL TWO-DOMAIN LECTIN | | Descriptor: | LECTIN SCAFET PRECURSOR | | Authors: | Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, VanDamme, E.J.M, Donovan, M.J, Peumans, W.J. | | Deposit date: | 1999-12-11 | | Release date: | 2000-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of the native fetuin-binding protein Scilla campanulata agglutinin: a novel two-domain lectin.
FEBS Lett., 468, 2000
|
|
8QID
 
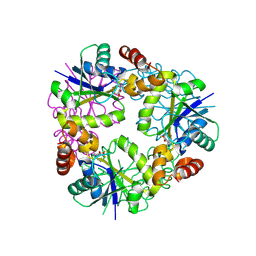 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with fragment | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QJ8
 
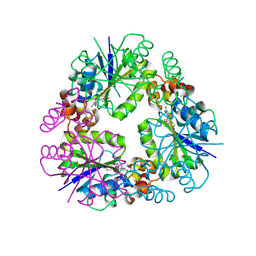 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 3-[3-(3-azanyl-2-cyano-phenyl)indol-1-yl]propanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QKS
 
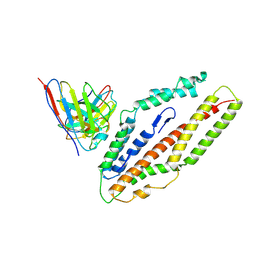 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.034 | | Descriptor: | Immunoglobulin lambda variable 1-36, R5034HV, Reticulocyte-binding protein-like protein 5 | | Authors: | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Von Delft, F, Koekemoer, L, Draper, S.J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.994 Å) | | Cite: | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
8QIY
 
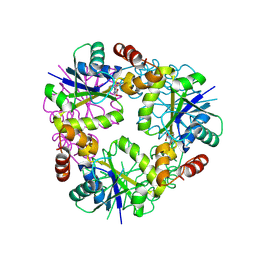 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 1-(2-aminophenyl)-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.5149 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QIX
 
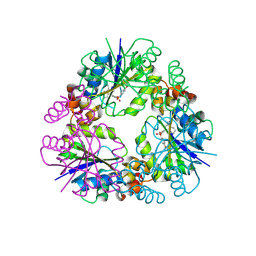 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 3-(3-methylindol-1-yl)-~{N}-(4-phenoxyphenyl)sulfonyl-propanamide, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QKR
 
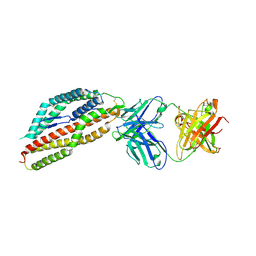 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.251 | | Descriptor: | R5251VHCH, R5251VLCL, Reticulocyte-binding protein-like protein 5 | | Authors: | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Koekemoer, L, Draper, S.J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.234 Å) | | Cite: | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
4EKF
 
 | |
3PMG
 
 | |
2TPT
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-24 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
2SAS
 
 | |
8K1Y
 
 | | Crystal structure of human serum albumin and ruthenium Val complex adduct | | Descriptor: | Albumin, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gong, W.J, Xie, L.L, Wang, W.M, Wang, H.F. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human serum albumin and ruthenium VaL complex adduct
To Be Published
|
|
1CB1
 
 | |
6X5P
 
 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 3-chloro-4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4XBF
 
 | | Structure of LSD1:CoREST in complex with ssRNA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Luka, Z, Loukachevitch, L.V, Martin, W.J, Wagner, C, Reiter, N.J. | | Deposit date: | 2014-12-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | G-quadruplex RNA binding and recognition by the lysine-specific histone demethylase-1 enzyme.
RNA, 22, 2016
|
|
6XTJ
 
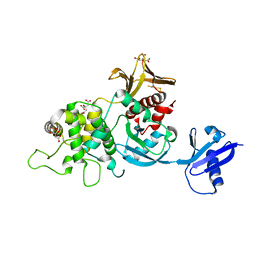 | | The high resolution structure of the FERM domain of human FERMT2 | | Descriptor: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
4XSQ
 
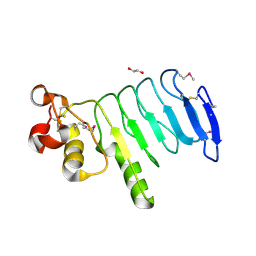 | | Structure of a variable lymphocyte receptor-like protein Bf66946 from Branchiostoma floridae | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, variable lymphocyte receptor-like protein Bf66946 | | Authors: | Cao, D.D, Cheng, W, Jiang, Y.L, Wang, W.J, Li, Q, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of a variable lymphocyte receptor-like protein from the amphioxus Branchiostoma floridae.
Sci Rep, 6, 2016
|
|
6XY7
 
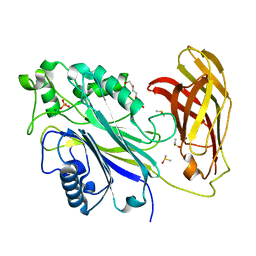 | | Human SHIP1 with magnesium and phosphate bound to the active site | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bradshaw, W.J, Scacioc, A, Fernandez-Cid, A, Mckinley, G, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-29 | | Release date: | 2020-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5I7M
 
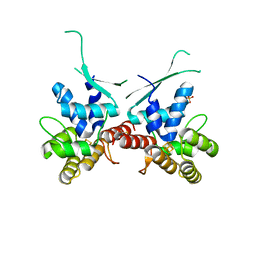 | |
6XQ1
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-[3-({[3-(4-carboxyphenoxy)phenyl]methoxy}methyl)phenyl]-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
1CNP
 
 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
6XQ6
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 11 | | Descriptor: | 3-phenoxyphenol, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ3
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-(3-{[3-(4-carboxyphenoxy)phenyl]methoxy}phenyl)-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ8
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 11 | | Descriptor: | 3-phenoxyphenol, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
