4W8K
 
 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | Descriptor: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
6U60
 
 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | Authors: | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
8K3D
 
 | | Crystal structure of NRF1 DBD bound to DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*CP*GP*CP*AP*TP*GP*CP*GP*CP*AP*CP*C)-3'), Nuclear respiratory factor 1 | | Authors: | Li, W.F, Liu, K, Min, J.R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of specific DNA sequence recognition by NRF1.
Nucleic Acids Res., 52, 2024
|
|
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
5O6B
 
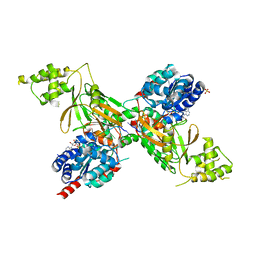 | | Structure of ScPif1 in complex with GGGTTTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(*GP*GP*GP*TP*TP*T)-3'), ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
5O6D
 
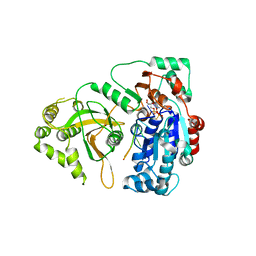 | | Structure of ScPif1 in complex with polydT and ATPgS | | Descriptor: | ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
5O6E
 
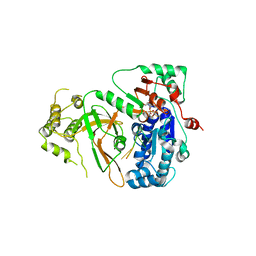 | | Structure of ScPif1 in complex with TTTGGGTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, CHLORIDE ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | Descriptor: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | Descriptor: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6ZDU
 
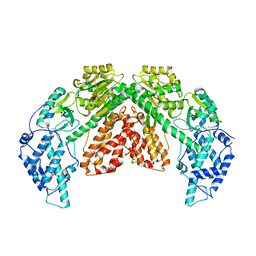 | | Structure of telomerase from Candida albicans in complexe with TWJ fragment of telomeric RNA | | Descriptor: | Chains: C,D, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZD2
 
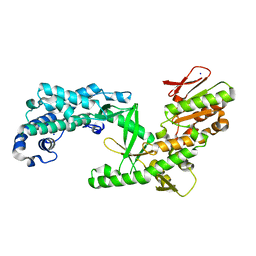 | | Structure of apo telomerase from Candida Tropicalis truncated from C-terminal domain | | Descriptor: | SODIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZDQ
 
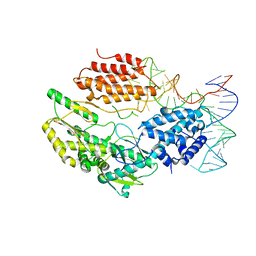 | | Structure of telomerase from Candida albicans in complexe with TWJ fragment of telomeric RNA | | Descriptor: | RNA, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZD1
 
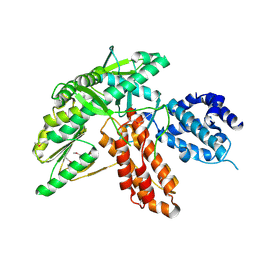 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZD6
 
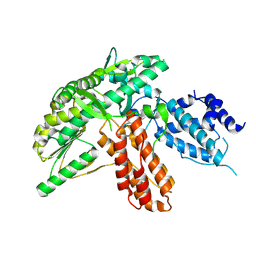 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZDP
 
 | | Structure of telomerase from Candida Tropicalis in complexe with TWJ fragment of telomeric RNA | | Descriptor: | Chains: B, POTASSIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6WMH
 
 | |
6WNA
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, Immunoglobulin heavy constant gamma 4, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WOL
 
 | | Next generation monomeric IgG4 Fc bound to neonatal Fc receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, IgG receptor FcRn large subunit p51, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6XZT
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai mutant G110C-E410C | | Descriptor: | PIF1 helicase | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.341 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
6YB1
 
 | | Crystal structure of an antiparallel octameric transmembrane coiled coil K2-CCTM-VbIc | | Descriptor: | GLYCINE, K2-CCTM-VbIc, NONAETHYLENE GLYCOL | | Authors: | Kratochvil, H.T, Liu, L, Scott, A.J, Woolfson, D.N, DeGrado, W.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Constructing ion channels from water-soluble alpha-helical barrels.
Nat.Chem., 13, 2021
|
|
8T3N
 
 | | Solution NMR structure of synthetic peptide AMPCry10Aa_5 rational designed from Cry10Aa bacterial protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | SOLUTION NMR | | Cite: | Anti-Staphy Peptides Rationally Designed from Cry10Aa Bacterial Protein.
Acs Omega, 9, 2024
|
|
8T3H
 
 | | Solution NMR structure alpha-helix 3 of Cry10Aa protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix 3 of Cry10Aa protein
To Be Published
|
|
1V45
 
 | | Crystal Structure of human PNP complexed with 3-deoxyguanosine | | Descriptor: | 9-(3-DEOXY-BETA-D-RIBOFURANOSYL)GUANINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Canduri, F, Dos Santos, D.M, Silva, R.G, Mendes, M.A, Palma, M.S, De Azevedo Jr, W.F, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-11-10 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8TNB
 
 | |
