3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | IODIDE ION, Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UUA
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with bisphenol-AF | | Descriptor: | 4,4'-(1,1,1,3,3,3-hexafluoropropane-2,2-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V70
 
 | | Crystal Structure of Human GTPase IMAP family member 1 | | Descriptor: | GTPase IMAP family member 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal Structure of Human GTPase IMAP family member 1
to be published
|
|
3V4E
 
 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | Authors: | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
3UEE
 
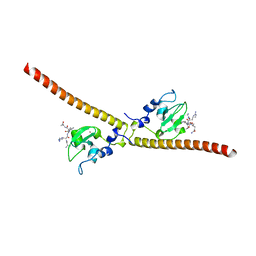 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
4K64
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus complexed with human receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4KAP
 
 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3UTM
 
 | | Crystal structure of a mouse Tankyrase-Axin complex | | Descriptor: | Axin-1, Tankyrase-1 | | Authors: | Cheng, Z, Morrone, S, Xu, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KGG
 
 | | Crystal structure of light mutant2 and dcr3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KGX
 
 | | The R state structure of E. coli ATCase with CTP bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
3V08
 
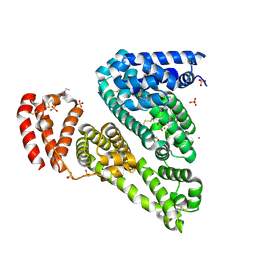 | | Crystal structure of Equine Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, SULFATE ION, ... | | Authors: | Dayal, A, Jablonska, K, Porebski, P.J, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
4KOB
 
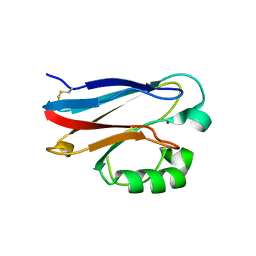 | |
4KOU
 
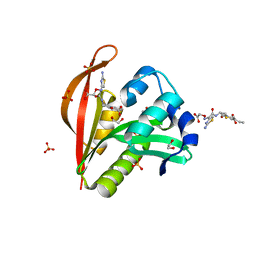 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefixime | | Descriptor: | (6R,7R)-7-({(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-[(carboxymethoxy)imino]acetyl}amino)-3-ethenyl-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KQE
 
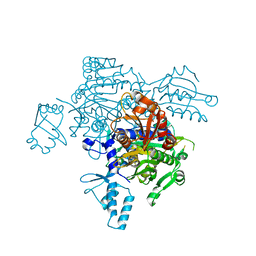 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
4KS0
 
 | | Pyruvate kinase (PYK) from Trypanosoma cruzi in the presence of Magnesium, oxalate and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Morgan, H.P, Zhong, W, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
4KRY
 
 | | Structure of Aes from E. coli in covalent complex with PMS | | Descriptor: | Acetyl esterase, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
3V5S
 
 | | Structure of Sodium/Calcium Exchanger from Methanococcus jannaschii | | Descriptor: | CADMIUM ION, Sodium/Calcium Exchanger | | Authors: | Liao, J, Li, H, Zeng, W, Sauer, D.B, Belmares, R, Jiang, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
4KE4
 
 | | Elucidation of the structure and reaction mechanism of Sorghum bicolor hydroxycinnamoyltransferase and its structural relationship to other CoA-dependent transferases and synthases | | Descriptor: | GLYCEROL, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase, TETRAETHYLENE GLYCOL | | Authors: | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
3UNB
 
 | | Mouse constitutive 20S proteasome in complex with PR-957 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UWN
 
 | | The 3-MBT repeat domain of L3MBTL1 in complex with a methyl-lysine mimic | | Descriptor: | Lethal(3)malignant brain tumor-like protein 1, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Wernimont, A.K, Graslund, S, Ingerman, L.A, Korboukh, V, Kireev, D.B, Gao, C, Frye, S.V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The 3-MBT repeat domain of L3MBTL1 in complex with a methyl-lysine mimic
To be Published
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
4L0G
 
 | | Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside hydrolase family 48, ... | | Authors: | Jiao, A, Bai, A, Yan, F, Geng, W. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary X-ray analysis of a processive cellobiohydrolase CbCBH48A from Caldicellulosiruptor bescii
to be published
|
|
4L1M
 
 | | Structure of the first RCC1-like domain of HERC2 | | Descriptor: | E3 ubiquitin-protein ligase HERC2, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Khan, M.B, Dong, A, Hu, J, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the first RCC1-like domain of HERC2
TO BE PUBLISHED
|
|
3V32
 
 | | Crystal structure of MCPIP1 N-terminal conserved domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
