3P8K
 
 | | Crystal Structure of a putative carbon-nitrogen family hydrolase from Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gordon, R.D, Qiu, W, Battaile, K, Lam, K, Soloveychik, M, Benetteraj, D, Romanov, V, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-10-14 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of carbon-nitrogen family hydrolase from Staphylococcus aureus
To be Published
|
|
5S73
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | Descriptor: | Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
4P0Z
 
 | |
5S74
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
1R2W
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of the 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
3OQN
 
 | | Structure of ccpa-hpr-ser46-p-gntr-down cre | | Descriptor: | 5'-D(*AP*TP*GP*GP*TP*AP*CP*CP*GP*CP*TP*TP*TP*CP*AP*A)-3', 5'-D(*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*TP*AP*CP*CP*AP*T)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
1R54
 
 | | Crystal structure of the catalytic domain of human ADAM33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reicher, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
4PBV
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform, ... | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
3NWN
 
 | | Crystal structure of the human KIF9 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF9, ... | | Authors: | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human KIF9 motor domain in complex with ADP
To be Published
|
|
4P0Y
 
 | |
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
5UP2
 
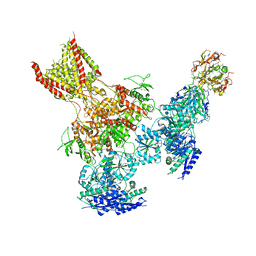 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, Ro 25-6981, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GluN2B-specific Fab, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
5URV
 
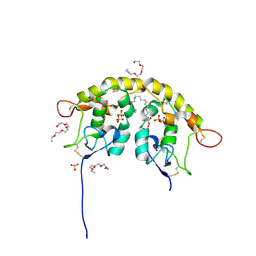 | | Crystal structure of Frizzled 7 CRD in complex with C24 fatty acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2PT6
 
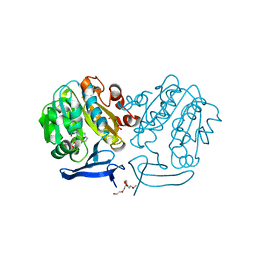 | | The structure of Plasmodium falciparum spermidine synthase in complex with decarboxylated S-adenosylmethionine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, GLYCEROL, ... | | Authors: | Dufe, V.T, Qiu, W, Muller, I.B, Hui, R, Walter, R.D, Al-Karadaghi, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.
J.Mol.Biol., 373, 2007
|
|
5URZ
 
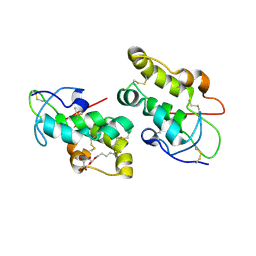 | | Crystal structure of Frizzled 5 CRD in complex with BOG | | Descriptor: | Frizzled-5, alpha-L-fucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, octyl beta-D-glucopyranoside | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2PSY
 
 | | Crystal Structure of Human Kallikrein 5 in complex with Leupeptin and Zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Kallikrein-5, LEUPEPTIN, ... | | Authors: | Debela, M, Bode, W, Goettig, P. | | Deposit date: | 2007-05-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the zinc inhibition of human tissue kallikrein 5.
J.Mol.Biol., 373, 2007
|
|
3O9X
 
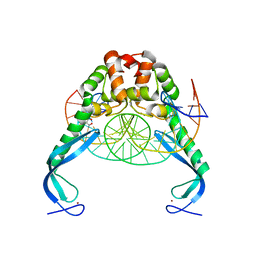 | | Structure of the E. coli antitoxin MqsA (YgiT/b3021) in complex with its gene promoter | | Descriptor: | DNA (26-MER), GLYCEROL, Uncharacterized HTH-type transcriptional regulator ygiT, ... | | Authors: | Brown, B.L, Peti, W, Page, R. | | Deposit date: | 2010-08-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.0999 Å) | | Cite: | Structure of the Escherichia coli Antitoxin MqsA (YgiT/b3021) Bound to Its Gene Promoter Reveals Extensive Domain Rearrangements and the Specificity of Transcriptional Regulation.
J.Biol.Chem., 286, 2011
|
|
3OE0
 
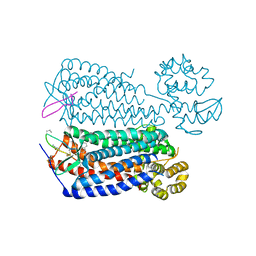 | | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 | | Descriptor: | C-X-C chemokine receptor type 4, Lysozyme Chimera, Polyphemusin analog, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
5ULC
 
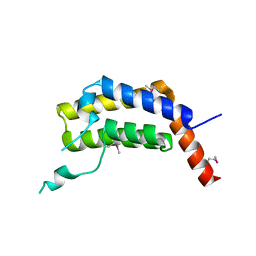 | | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328 | | Descriptor: | Bromodomain protein 1 | | Authors: | Wernimont, A.K, Amaya, M.F, Lam, A, Ali, A, Zhang, A.Z, Kenzina, L, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Hui, R, Walker, J.R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328
To be published
|
|
2PWP
 
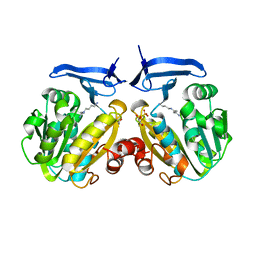 | | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermidine | | Descriptor: | GLYCEROL, SPERMIDINE, SULFATE ION, ... | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Zhao, Y, Schapira, M, Wasney, G, Vedadi, M, Lew, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of spermidine synthase from Plasmodium falciparum in complex with spermidine.
To be Published
|
|
1UY7
 
 | | Human Hsp90-alpha with 9-Butyl-8-(4-methoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(4-METHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
7CRR
 
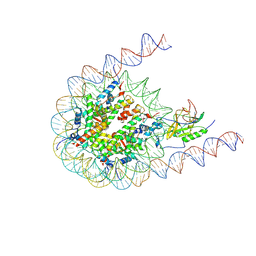 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
2PFL
 
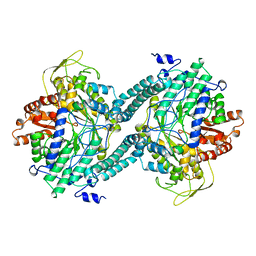 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | Descriptor: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-26 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1URS
 
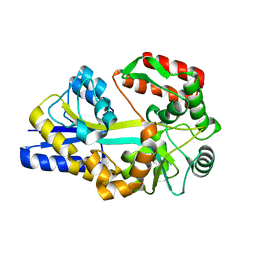 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1UYC
 
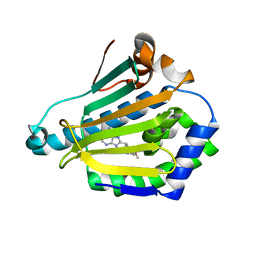 | | Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2,5-DIMETHOXY-BENZYL)-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
