4G5R
 
 | | Structure of LGN GL4/Galphai3 complex | | 分子名称: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | 登録日 | 2012-07-18 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.481 Å) | | 主引用文献 | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | 分子名称: | ELASTASE, SODIUM ION, SULFATE ION | | 著者 | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | 登録日 | 1999-10-15 | | 公開日 | 2000-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | 分子名称: | Isocitrate dehydrogenase [NADP], SULFATE ION | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-11-23 | | 公開日 | 2011-12-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3UUG
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | 分子名称: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | 著者 | Hu, X, Zhao, J, Binns, A, Degrado, W. | | 登録日 | 2011-11-28 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3URM
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | 分子名称: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | 著者 | Hu, X, Zhao, J, Binns, A, Degrado, W. | | 登録日 | 2011-11-22 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UXY
 
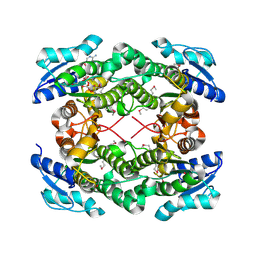 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | 著者 | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-05 | | 公開日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
2SGP
 
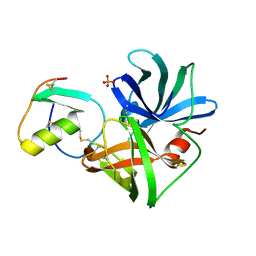 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | 分子名称: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2001-01-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
8BTM
 
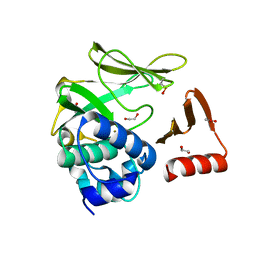 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | 著者 | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | 登録日 | 2022-11-29 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
3V2G
 
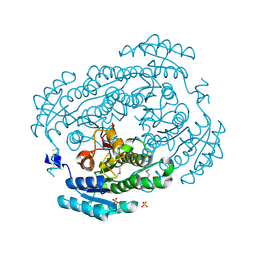 | | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-12 | | 公開日 | 2012-01-04 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4G5O
 
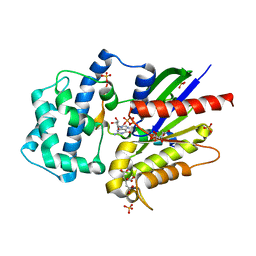 | | Structure of LGN GL4/Galphai3(Q147L) complex | | 分子名称: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | 登録日 | 2012-07-18 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
8CF9
 
 | | Crystal structure of the human PXR ligand-binding domain in complex with sclareol | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, sclareol | | 著者 | Carivenc, C, Derosa, Q, Grimaldi, M, Boulahtouf, A, Balaguer, P, Bourguet, W. | | 登録日 | 2023-02-03 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the human PXR ligand-binding domain in complex with sclareol
To Be Published
|
|
8CH8
 
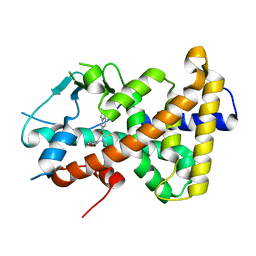 | | Crystal structure of the human PXR ligand-binding domain in complex with liranaftate | | 分子名称: | Nuclear receptor subfamily 1 group I member 2, ~{O}-(5,6,7,8-tetrahydronaphthalen-2-yl) ~{N}-(6-methoxypyridin-2-yl)-~{N}-methyl-carbamothioate | | 著者 | Carivenc, C, Derosa, Q, Grimaldi, M, Boulahtouf, A, Balaguer, P, Bourguet, W. | | 登録日 | 2023-02-07 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of the human PXR ligand-binding domain in complex with liranaftate
To Be Published
|
|
8F3H
 
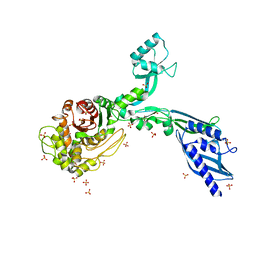 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3L
 
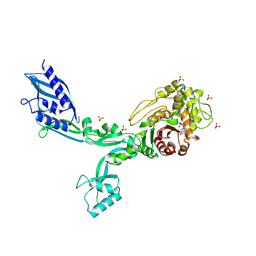 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3O
 
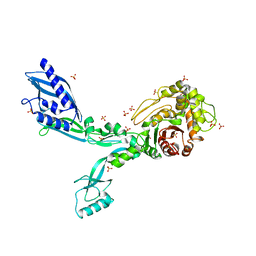 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Peti, W, Page, R. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3S
 
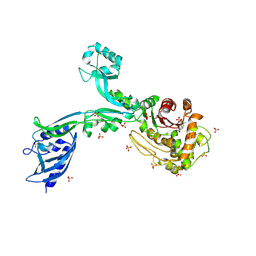 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F67
 
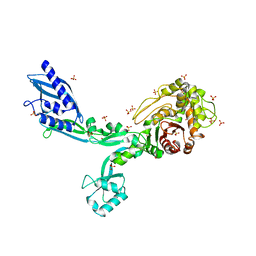 | | Crystal structure of the refolded Penicillin Binding Protein 5 (PBP5) of Enterococcus faecium | | 分子名称: | Pbp5, SULFATE ION | | 著者 | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Peti, W, Page, R. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3T
 
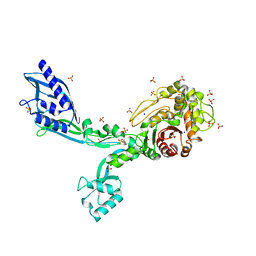 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3I
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Z
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S422A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3U
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3N
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3F
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Hunashal, Y, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3R
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3P
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
