4TNZ
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dATP-dTTP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OWA
 
 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
3OSI
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with tetrachloro-bisphenol A (TCBPA) | | Descriptor: | 4,4'-propane-2,2-diylbis(2,6-dichlorophenol), Peroxisome proliferator-activated receptor gamma, S-1,2-PROPANEDIOL | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2010-09-09 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peroxisome proliferator-activated receptor Gamma is a target for halogenated analogs of bisphenol A.
Environ.Health Perspect., 119, 2011
|
|
3OUG
 
 | | Crystal structure of cleaved L-aspartate-alpha-decarboxylase from Francisella tularensis | | Descriptor: | Aspartate 1-decarboxylase, CHLORIDE ION, GLYCEROL | | Authors: | Nocek, B, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of cleaved L-aspartate-alpha-decarboxylase from Francisella tularensis
To be Published
|
|
4TTC
 
 | |
4TKM
 
 | | Crystal structure of NADH-dependent reductase A1-R' complexed with NAD | | Descriptor: | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
5TEY
 
 | | Human METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | DONG, A, ZENG, H, LI, Y, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-23 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human METTL3-METTL14 complex
to be published
|
|
4TNR
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dATP-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TO4
 
 | | Structure basis of cellular dNTP regulation, SAMHD1-GTP-dGTP-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Ji, X, Tang, C, Zhao, Q, Wang, W, Xiong, Y. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cellular dNTP regulation by SAMHD1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TS0
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, barium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, BARIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7DUR
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
3LIJ
 
 | | Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP | | Descriptor: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, MAGNESIUM ION, ... | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-25 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP
To be Published
|
|
5TIG
 
 | |
5UNF
 
 | | XFEL structure of human angiotensin II type 2 receptor (Monoclinic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]) | | Descriptor: | Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
4RLZ
 
 | | Crystal structure of Norovirus OIF P domain | | Descriptor: | Capsid protein, GLYCEROL | | Authors: | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
2PAU
 
 | | Crystal structure of the 5'-deoxynucleotidase YfbR mutant E72A complexed with Co(2+) and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-deoxynucleotidase YfbR, COBALT (II) ION, ... | | Authors: | Zimmerman, M.D, Proudfoot, M, Yakunin, A, Minor, W. | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
J.Mol.Biol., 378, 2008
|
|
5UOG
 
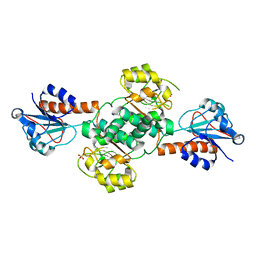 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in apo form | | Descriptor: | NADPH-dependent glyoxylate/hydroxypyruvate reductase, SULFATE ION | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
4RMJ
 
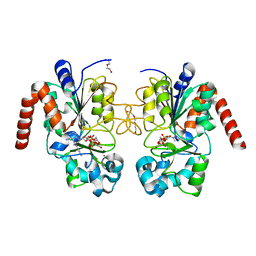 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMH
 
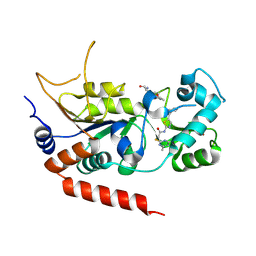 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
3LOS
 
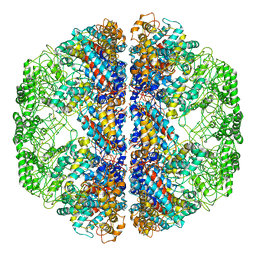 | | Atomic Model of Mm-cpn in the Closed State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
4RSY
 
 | | Crystal structures of the Human leukotriene A4 Hydrolase complex with a potential inhibitor H7 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, ACETIC ACID, IMIDAZOLE, ... | | Authors: | Ouyang, P, Lu, W, Cui, K, Huang, J. | | Deposit date: | 2014-11-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of Human Leukotriene A4 Hydrolase in complex with inhibitor H1
To be Published
|
|
4ROF
 
 | | Crystal Structure of WW3 domain of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of WW3 domain of ITCH in complex with TXNIP peptide
To be Published
|
|
3R4F
 
 | | Prohead RNA | | Descriptor: | MAGNESIUM ION, pRNA | | Authors: | Ding, F, Lu, C, Zhano, W, Rajashankar, K.R, Anderson, D.L, Jardine, P.J, Grimes, S, Ke, A. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and assembly of the essential RNA ring component of a viral DNA packaging motor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5UVK
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
Iucrj, 4, 2017
|
|
5UX3
 
 | |
