6MT7
 
 | |
6MTF
 
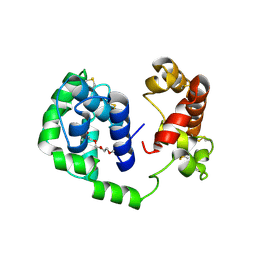 | | D7 protein from Phlebotomus duboscqi, native | | Descriptor: | 26.7 kDa salivary protein, FRAGMENT OF TRITON X-100 | | Authors: | Andersen, J.F, Jablonka, W. | | Deposit date: | 2018-10-19 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional and structural similarities of D7 proteins in the independently-evolved salivary secretions of sand flies and mosquitoes.
Sci Rep, 9, 2019
|
|
6MH6
 
 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6MKA
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the open conformation | | Descriptor: | SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
1W77
 
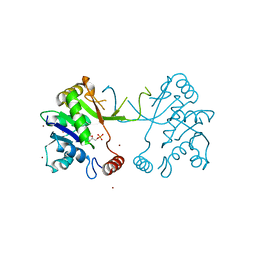 | | 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD) from Arabidopsis thaliana | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CADMIUM ION, COPPER (II) ION, ... | | Authors: | Gabrielsen, M, Kaiser, J, Rohdich, F, Eisenreich, W, Bacher, A, Bond, C.S, Hunter, W.N. | | Deposit date: | 2004-08-30 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Plant 2C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase Exhibits a Distinct Quaternary Structure Compared to Bacterial Homologues and a Possible Role in Feedback Regulation for Cytidine Monophosphate.
FEBS J., 273, 2006
|
|
1W4Z
 
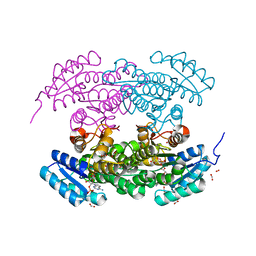 | | Structure of actinorhodin polyketide (actIII) Reductase | | Descriptor: | FORMIC ACID, KETOACYL REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Limpkin, C, Teartasin, W, Simpson, T.J, Crosby, J, Crump, M.P. | | Deposit date: | 2004-08-03 | | Release date: | 2004-12-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the Actiii Actinorhodin Polyketide Reductase; Proposed Mechanism for Acp and Polyketide Binding
Structure, 12, 2004
|
|
6MKG
 
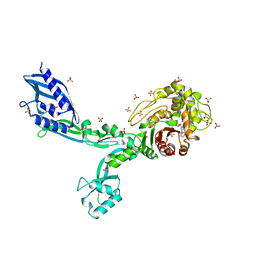 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the benzylpenicilin-bound form | | Descriptor: | OPEN FORM - PENICILLIN G, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MJW
 
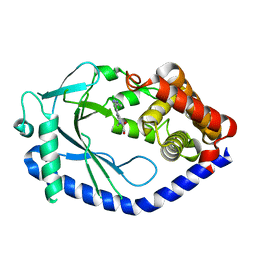 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
4D4F
 
 | | Mutant P250A of bacterial chalcone isomerase from Eubacterium ramulus | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL | | Authors: | Thomsen, M, Kratzat, H, Hinrichs, W. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus.
Molecules, 27, 2022
|
|
4BWE
 
 | | Crystal structure of C-terminally truncated glypican-1 after controlled dehydration to 86 percent relative humidity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glypican-1 | | Authors: | Awad, W, Svensson Birkedal, G, Thunnissen, M.M.G.M, Mani, K, Logan, D.T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Improvements in the order, isotropy and electron density of glypican-1 crystals by controlled dehydration.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
1VYD
 
 | | Crystal structure of cytochrome C2 mutant G95E | | Descriptor: | CYTOCHROME C2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dumortier, C, Fitch, J, Van Petegem, F, Vermeulen, W, Meyer, T.E, Van Beeumen, J.J, Cusanovich, M.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein Dynamics in the Region of the Sixth Ligand Methionine Revealed by Studies of Imidazole Binding to Rhodobacter Capsulatus Cytochrome C2 Hinge Mutants.
Biochemistry, 43, 2004
|
|
6N31
 
 | | WD repeats of human WDR12 | | Descriptor: | Ribosome biogenesis protein WDR12, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Tempel, W, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | WD repeats of human WDR12
To Be Published
|
|
1YQL
 
 | | Catalytically inactive hOGG1 crosslinked with 7-deaza-8-azaguanine containing DNA | | Descriptor: | 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*CP*AP*(PPW)P*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
1YQ9
 
 | | Structure of the unready oxidized form of [NiFe] hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
6MRW
 
 | | 14-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6N7W
 
 | | Structure of bacteriophage T7 leading-strand DNA polymerase (D5A/E7A)/Trx in complex with a DNA fork and incoming dTTP (from multiple lead complexes) | | Descriptor: | DNA (25-MER), DNA (77-MER), DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6MIW
 
 | | WWE domain of human HUWE1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, E3 ubiquitin-protein ligase HUWE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Loppnau, P, Tempel, W, Wong, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WWE domain of human HUWE1
To Be Published
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
1XQ4
 
 | | Crystal Structure of the Putative ApaA Protein from Bordetella pertussis, Northeast Structural Genomics Target BeR40 | | Descriptor: | PHOSPHATE ION, Protein apaG | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative ApaA Protein from Bordetella pertussis, Northeast Structural Genomics Target BeR40
To be Published
|
|
1XRP
 
 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Pro-Leu-Gly-Gly | | Descriptor: | PLGG, PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
4GK9
 
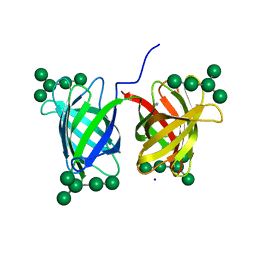 | | Crystal structure of Burkholderia oklahomensis agglutinin (BOA) bound to 3a,6a-mannopentaose | | Descriptor: | IMIDAZOLE, SODIUM ION, agglutinin (BOA), ... | | Authors: | Whitley, M.J, Furey, W, Gronenborn, A.M. | | Deposit date: | 2012-08-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: structures, carbohydrate binding and anti-HIV activity.
Febs J., 280, 2013
|
|
1XI6
 
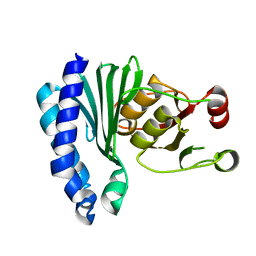 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | Descriptor: | extragenic suppressor | | Authors: | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
6N7I
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (gp4(5)-DNA) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N9U
 
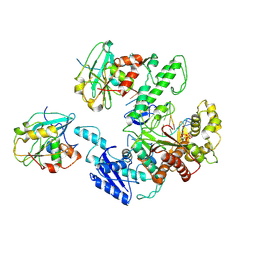 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) interacting with primase domains of two gp4 subunits bound to an RNA/DNA hybrid and dTTP (from LagS1) | | Descriptor: | DNA (44-MER), DNA primase/helicase, DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
