6XTY
 
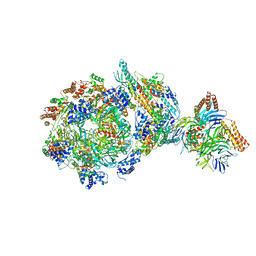 | | CryoEM structure of human CMG bound to AND-1 (CMGA) | | Descriptor: | Cell division control protein 45 homolog, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Rzechorzek, N.J, Pellegrini, L, Chirgadze, D.Y, Hardwick, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
7NKF
 
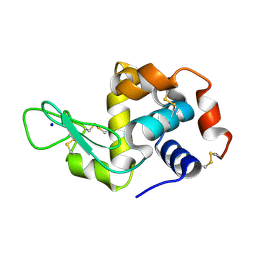 | | Hen egg white lysozyme (HEWL) Grown inside (Not centrifuged) HARE serial crystallography chip. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJE
 
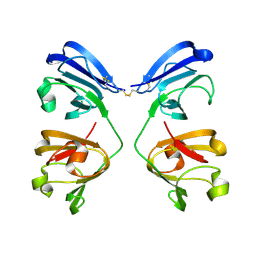 | | gamma(S)-crystallin 9-site deamidation mutant grown inside HARE serial crystallography chip | | Descriptor: | Gamma-crystallin S | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJH
 
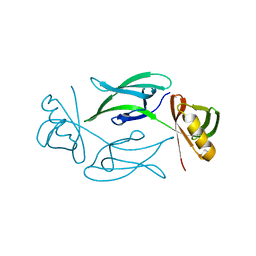 | | HEX1 (in cellulo) grown inside HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6XYO
 
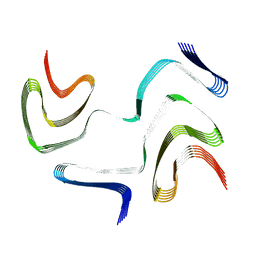 | | Multiple system atrophy Type I alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
7N63
 
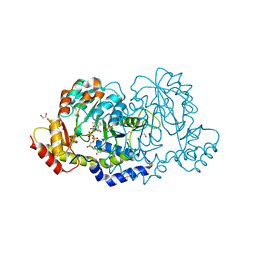 | | X-ray structure of HCAN_0200, an aminotransferase from Helicobacter canadensis in complex with its external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Griffiths, W.A, Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7NJI
 
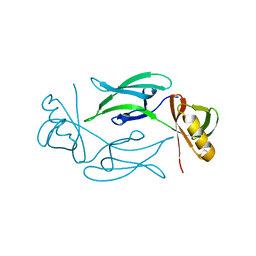 | | HEX1 (in cellulo) loaded on HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DHQ
 
 | | Structure of Halothiobacillus neapolitanus Microcompartments Protein CsoS1D | | Descriptor: | Microcompartments protein | | Authors: | Xue, B, Tan, Y.Q, Ali, S, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7N8C
 
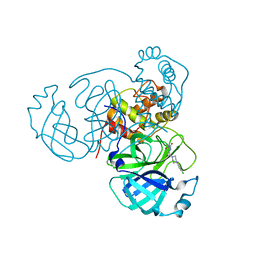 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-06-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7N89
 
 | |
7N67
 
 | | Crystal structure of HCAN_0198, a 3,4-ketoisomerase from Helicobacter canadensis | | Descriptor: | FdtA domain-containing protein, TETRAMETHYLAMMONIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Heisdorf, C.J, Griffiths, W.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
3QBY
 
 | | Crystal structure of the PWWP domain of human Hepatoma-derived growth factor 2 | | Descriptor: | H4K20me3 Histone H4 Peptide, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Zeng, H, Tempel, W, Amaya, M.F, Adams-Cioaba, M.A, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
6XHU
 
 | |
1S8O
 
 | | Human soluble Epoxide Hydrolase | | Descriptor: | HEXAETHYLENE GLYCOL, epoxide hydrolase 2, cytoplasmic | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
6X94
 
 | | An orthogonal seryl-tRNA synthetase/tRNA pair for noncanonical amino acid mutagenesis in Escherichia coli | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zambaldo, C, Koh, M, Nasertorabi, F, Han, G.W, Chatterjee, A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An orthogonal seryl-tRNA synthetase/tRNA pair for noncanonical amino acid mutagenesis in Escherichia coli.
Bioorg.Med.Chem., 28, 2020
|
|
7NG2
 
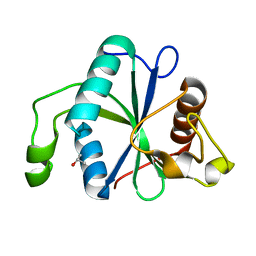 | | Crystal structure of Toxoplasma CPSF4-YTH domain in apo form | | Descriptor: | ISOPROPYL ALCOHOL, Zinc finger (CCCH type) motif-containing protein | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
7NJC
 
 | |
8BOU
 
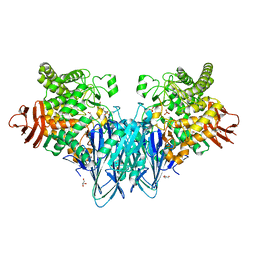 | | Crystal structure of Blautia producta GH94 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Blautia producta is a competent degrader among human gut Firmicutes for utilizing dietary beta mixed linkage glucan
To Be Published
|
|
1SOJ
 
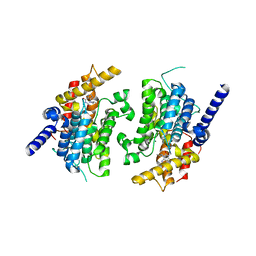 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B IN COMPLEX WITH IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, cGMP-inhibited 3',5'-cyclic phosphodiesterase B | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
7NH2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain bound to m6A | | Descriptor: | TETRAETHYLENE GLYCOL, Zinc finger (CCCH type) motif-containing protein, ~{N},9-dimethylpurin-6-amine | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
4OC9
 
 | | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205 | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205
To be Published
|
|
7N44
 
 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
6XNT
 
 | | Crystal structure of I91A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
1SL6
 
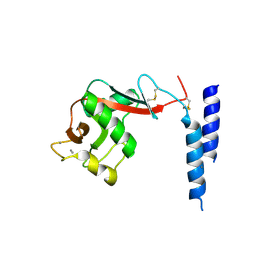 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
8GZE
 
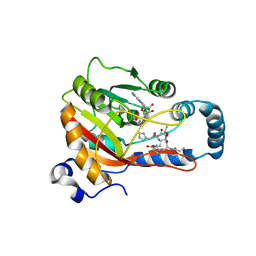 | | Crystal Structure of human METTL9-SAH-SLC39A7 peptide complex | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, Zinc transporter SLC39A7 | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
