6WV8
 
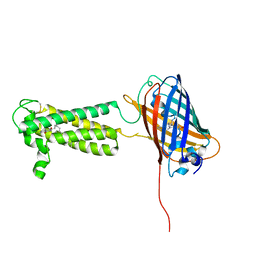 | | Takifugu rubripes VKOR-like C138S mutant with vitamin K1 | | Descriptor: | PHYLLOQUINONE, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
4AYX
 
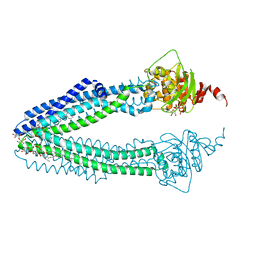 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (ROD FORM B) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4UFT
 
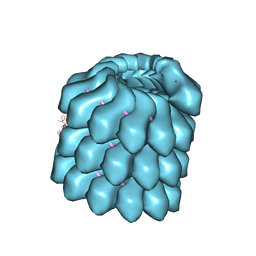 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
4BBB
 
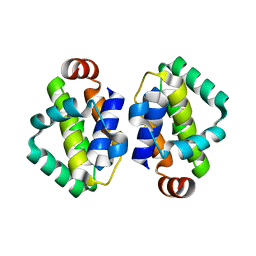 | | THE STRUCTURE OF VACCINIA VIRUS N1 Q61Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
4UPB
 
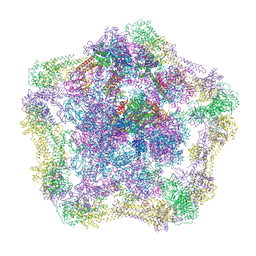 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
4BBD
 
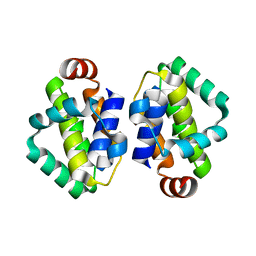 | | THE STRUCTURE OF VACCINIA VIRUS N1 R58Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
6WOZ
 
 | |
6WTY
 
 | |
8S32
 
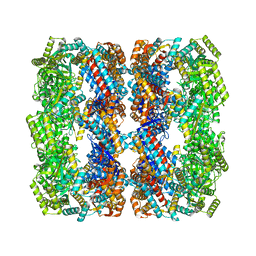 | | GroEL with bound GroTAC peptide | | Descriptor: | Chaperonin GroEL, GroTAC | | Authors: | Wroblewski, K, Izert-Nowakowska, M.A, Goral, T.K, Klimecka, M.M, Kmiecik, S, Gorna, M.W. | | Deposit date: | 2024-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Depletion of essential GroEL protein in Escherichia coli using Clp-Interacting Peptidic Protein Erasers (CLIPPERs)
To Be Published
|
|
4B8J
 
 | | rImp_alpha1a | | Descriptor: | IMPORTIN SUBUNIT ALPHA-1A | | Authors: | Chang, C.-W, Counago, R.L.M, Williams, S.J, Boden, M, Kobe, B. | | Deposit date: | 2012-08-28 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Rice Importin-Alpha and Structural Basis of its Interaction with Plant-Specific Nuclear Localization Signals.
Plant Cell, 24, 2012
|
|
4UJ6
 
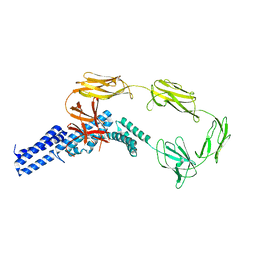 | | Structure of surface layer protein SbsC, domains 1-6 | | Descriptor: | SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 1-6
To be Published
|
|
4BB0
 
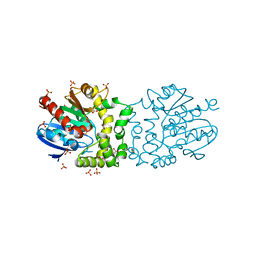 | |
7PHY
 
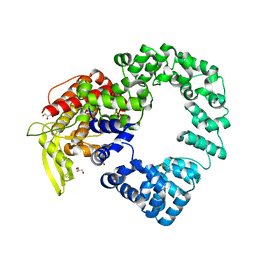 | | Vaccinia virus E2 | | Descriptor: | GLYCEROL, Protein E2 | | Authors: | Gao, W.N.D, Gao, C, Graham, S.C. | | Deposit date: | 2021-08-19 | | Release date: | 2021-09-01 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of vaccinia virus protein E2 and perspectives on the prediction of novel viral protein folds.
J.Gen.Virol., 103, 2022
|
|
7CF6
 
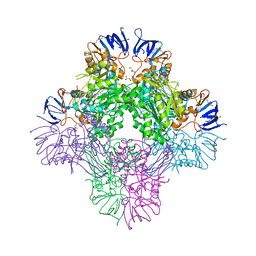 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
4B7K
 
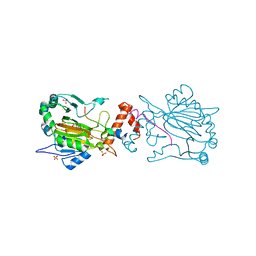 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4UID
 
 | |
4B4W
 
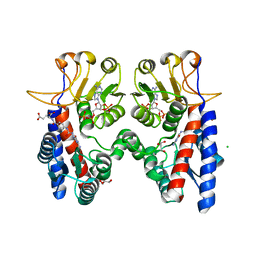 | | Crystal structure of Acinetobacter baumannii N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and an inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, BIFUNCTIONAL PROTEIN FOLD, CHLORIDE ION, ... | | Authors: | Eadsforth, T.C, Maluf, F.V, Hunter, W.N. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acinetobacter Baumannii Fold Ligand Complexes; Potent Inhibitors of Folate Metabolism and a Re-Evaluation of the Ly374571 Structure.
FEBS J., 279, 2012
|
|
4B7E
 
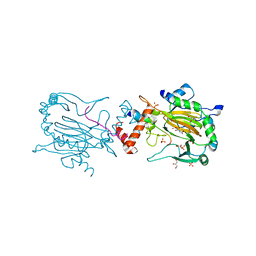 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-LEU PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-LEU, GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4B9P
 
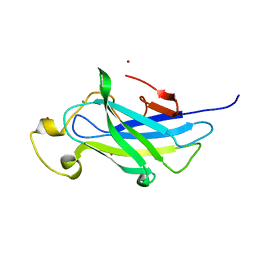 | | Biomass sensoring module from putative Rsgi2 protein of Clostridium thermocellum resemble family 3 carbohydrate-binding module of cellulosome | | Descriptor: | CALCIUM ION, TYPE 3A CELLULOSE-BINDING DOMAIN PROTEIN, ZINC ION | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Fine-Structural Variance of Family 3 Carbohydrate-Binding Modules as Extracellular Biomass-Sensing Components of Clostridium Thermocellum Anti-Sigma(I) Factors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UEG
 
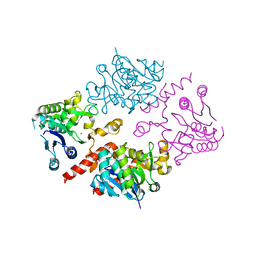 | | Crystal structure of human glycogenin-2 catalytic domain | | Descriptor: | GLYCOGENIN-2, MAGNESIUM ION | | Authors: | Fairhead, M, Strain-Damerell, C, Krojer, T, Froese, D.S, Kopec, J, Nowak, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Human Glycogenin-2 Catalytic Domain
To be Published
|
|
4BBA
 
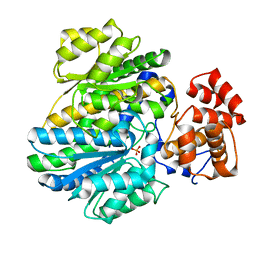 | | Crystal structure of glucokinase regulatory protein complexed to phosphate | | Descriptor: | GLUCOKINASE REGULATORY PROTEIN, PHOSPHATE ION | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinhart, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
4BBC
 
 | | THE STRUCTURE OF VACCINIA VIRUS N1 R71Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
4BKX
 
 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
6WO6
 
 | |
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
