1NWO
 
 | |
3IJW
 
 | | Crystal structure of BA2930 in complex with CoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
5F5Z
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MA2-014 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-~{N}-[3-[[5-methyl-2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]amino]phenyl]propane-2-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
5Y5V
 
 | |
8TMS
 
 | | Crystal structure of bacterial pectin methylesterase PmeC2 from rumen Butyrivibrio | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pectinesterase | | Authors: | Carbone, V, Reilly, K, Sang, C, Schofield, L, Ronimus, R, Kelly, W.J, Attwood, G.T, Palevich, N. | | Deposit date: | 2023-07-30 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Bacterial Pectin Methylesterases Pme8A and PmeC2 from Rumen Butyrivibrio .
Int J Mol Sci, 24, 2023
|
|
5F60
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SG3-014 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
3IOS
 
 | | Structure of MTB dsbF in its mixed oxidized and reduced forms | | Descriptor: | Disulfide bond forming protein (DsbF) | | Authors: | Chim, N, Goulding, C.W. | | Deposit date: | 2009-08-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An extracellular disulfide bond forming protein (DsbF) from Mycobacterium tuberculosis: structural, biochemical, and gene expression analysis.
J.Mol.Biol., 396, 2010
|
|
6YOS
 
 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pT524 pS617 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Glucocorticoid receptor,Glucocorticoid receptor | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
3IRX
 
 | | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the Non-nucleoside RT Inhibitor (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate. | | Descriptor: | (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methy lbenzothioate, Reverse transcriptase, Reverse transcriptase/ribonuclease H | | Authors: | Ho, W.C, Arnold, E. | | Deposit date: | 2009-08-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the
alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor (E)-S-Methyl
5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate.
J.Med.Chem., 52, 2009
|
|
3IOT
 
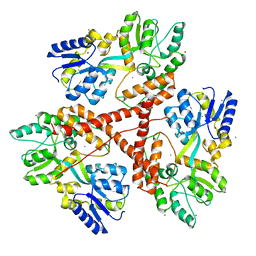 | |
8TAR
 
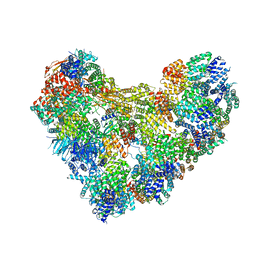 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
1OVO
 
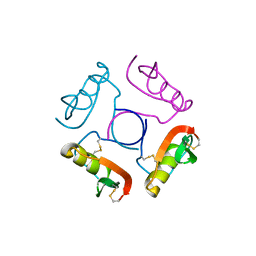 | | CRYSTALLOGRAPHIC REFINEMENT OF JAPANESE QUAIL OVOMUCOID, A KAZAL-TYPE INHIBITOR, AND MODEL BUILDING STUDIES OF COMPLEXES WITH SERINE PROTEASES | | Descriptor: | OVOMUCOID THIRD DOMAIN | | Authors: | Weber, E, Papamokos, E, Bode, W, Huber, R, Kato, I, Laskowskijunior, M. | | Deposit date: | 1982-01-18 | | Release date: | 1982-05-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic refinement of Japanese quail ovomucoid, a Kazal-type inhibitor, and model building studies of complexes with serine proteases.
J.Mol.Biol., 158, 1982
|
|
8TAU
 
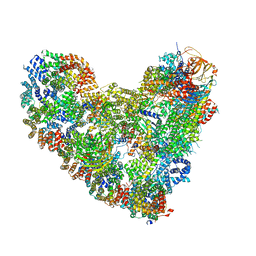 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
2CHB
 
 | | CHOLERA TOXIN B-PENTAMER COMPLEXED WITH GM1 PENTASACCHARIDE | | Descriptor: | CHOLERA TOXIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 1997-06-03 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of receptor binding by cholera toxin mutants.
Protein Sci., 6, 1997
|
|
5WQ0
 
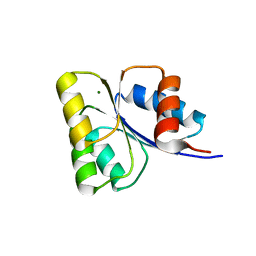 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
2CHT
 
 | |
5WYQ
 
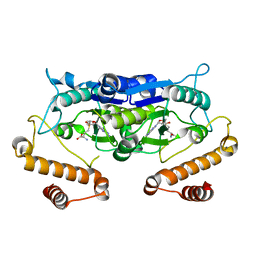 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
2CDO
 
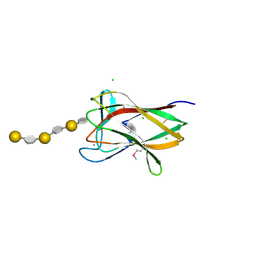 | | structure of agarase carbohydrate binding module in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-25 | | Release date: | 2006-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
4C3X
 
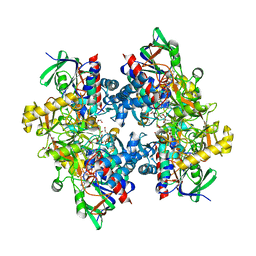 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
3IK7
 
 | | Human glutathione transferase a4-4 with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A4, SULFATE ION | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
1ETS
 
 | |
4QDS
 
 | | Physical basis for Nrp2 ligand binding | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-2 | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
6BAJ
 
 | |
2C9F
 
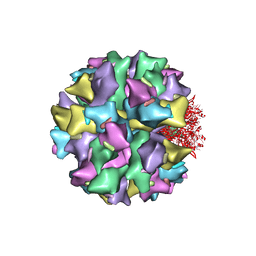 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON DODECAHEDRON | | Descriptor: | FIBER, PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
3DXA
 
 | | Crystal Structure of the DM1 TCR in complex with HLA-B*4405 and decamer EBV antigen | | Descriptor: | Beta-2-microglobulin, DM1 T cell receptor alpha chain, DM1 T cell receptor beta chain, ... | | Authors: | Archbold, J.K, Macdonald, W.A, Gras, S, Rossjohn, J. | | Deposit date: | 2008-07-23 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
J.Exp.Med., 206, 2009
|
|
