7EW4
 
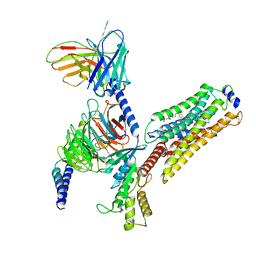 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
3UOI
 
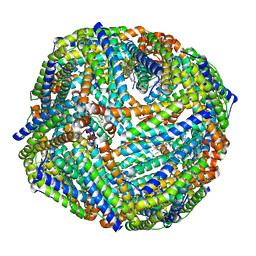 | |
3UF0
 
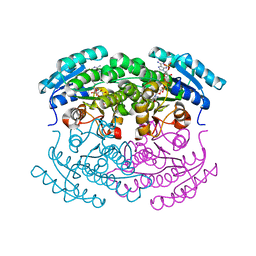 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
7VNG
 
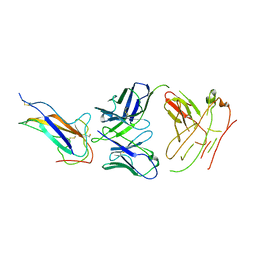 | | Crystal structure of human coronavirus 229E spike protein receptor-binding domain in complex with S11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S11 Fab heavy chain, S11 Fab light chain, ... | | Authors: | Xiang, J.C, Zhao, W.W, Yang, B. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Antigenic mapping reveals sites of vulnerability on alpha-HCoV spike protein.
Commun Biol, 5, 2022
|
|
3UGH
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
7EVZ
 
 | | Cryo-EM structure of cenerimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | (2~{S})-3-[4-[5-(2-cyclopentyl-6-methoxy-pyridin-4-yl)-1,2,4-oxadiazol-3-yl]-2-ethyl-6-methyl-phenoxy]propane-1,2-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
6HQL
 
 | | Crystal structure of GcoA F169H bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1FPG
 
 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
8TK7
 
 | |
2WE0
 
 | | EBV dUTPase mutant Cys4Ser | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, MALATE LIKE INTERMEDIATE, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
7EVY
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
6HQT
 
 | | Crystal structure of GcoA F169V bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8TWU
 
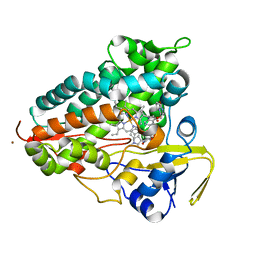 | | Crystal structure of Cytochrome P450 AspB bound to N1-methylated cyclo-L-Trp-L-Pro | | Descriptor: | (3S,5S,8aS)-3-[(1-methyl-1H-indol-3-yl)methyl]hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450 AspB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gering, H.E, Li, X, Tang, H, Swartz, P.D, Chang, W.-C, Makris, T.M. | | Deposit date: | 2023-08-21 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Ferric-Superoxide Intermediate Initiates P450-Catalyzed Cyclic Dipeptide Dimerization.
J.Am.Chem.Soc., 145, 2023
|
|
7EW3
 
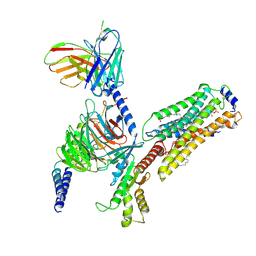 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
3UV7
 
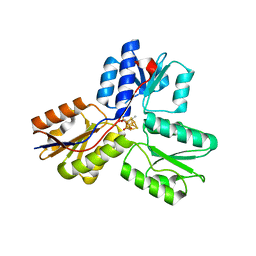 | | Ec_IspH in complex with buta-2,3-dienyl diphosphate (1300) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, buta-2,3-dien-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
1PJ1
 
 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F SOAKED WITH FERROUS IONS AT PH 5 | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
2NP1
 
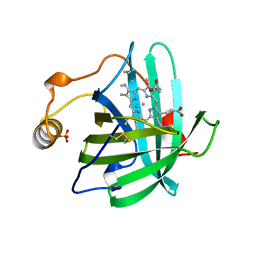 | |
1FK0
 
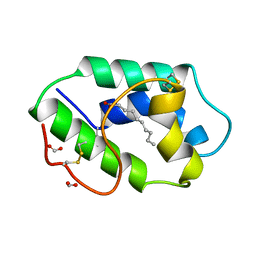 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
8S9V
 
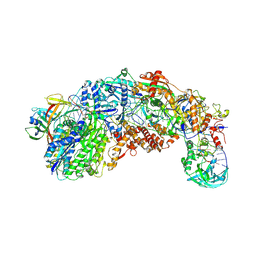 | |
8S9X
 
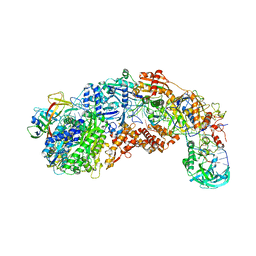 | |
1FGO
 
 | |
3PPQ
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | CHOLINE ION, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
4CIN
 
 | | Complex of Bcl-xL with its BH3 domain | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D. | | Deposit date: | 2013-12-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
J.Biol.Chem., 289, 2014
|
|
7ESC
 
 | | FmnB complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
