1C1M
 
 | | PORCINE ELASTASE UNDER XENON PRESSURE (8 BAR) | | Descriptor: | CALCIUM ION, PROTEIN (PORCINE ELASTASE), SULFATE ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
4NMU
 
 | | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor'
To be Published
|
|
8T6R
 
 | |
7EMD
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-GLY-ASP-PHE-PHE-HIS-ASP-MET-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
8PDI
 
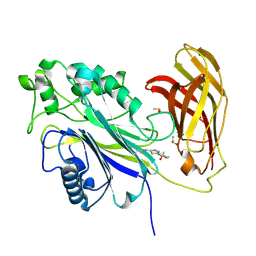 | | The phosphatase and C2 domains of SHIP1 with covalent Z1763271112 | | Descriptor: | (5-phenyl-1,3,4-thiadiazol-2-yl)methanimine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
7E2H
 
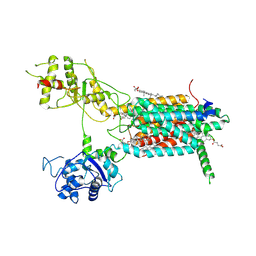 | | Cryo-EM structure of hDisp1NNN-3C-Cleavage | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
3UV6
 
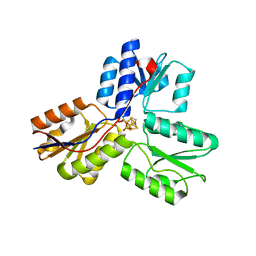 | | Ec_IspH in complex with 4-hydroxybutyl diphosphate (1301) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-hydroxybutyl trihydrogen diphosphate, IRON/SULFUR CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1BTG
 
 | |
3PBL
 
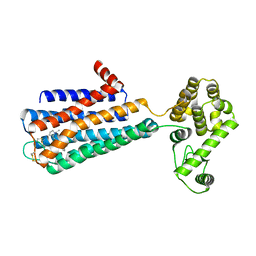 | | Structure of the human dopamine D3 receptor in complex with eticlopride | | Descriptor: | 3-chloro-5-ethyl-N-{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxy-2-methoxybenzamide, D(3) dopamine receptor, Lysozyme chimera, ... | | Authors: | Chien, E.Y.T, Liu, W, Han, G.W, Katritch, V, Zhao, Q, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the human dopamine d3 receptor in complex with a d2/d3 selective antagonist.
Science, 330, 2010
|
|
7WPP
 
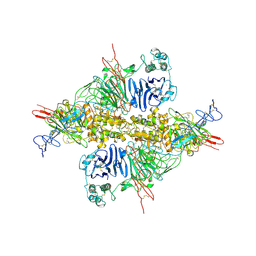 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 2.85 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
3UWM
 
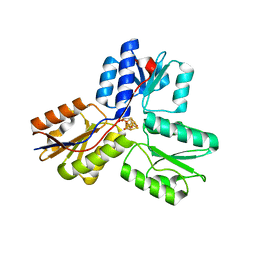 | | Ec_IspH in complex with 4-oxobutyl diphosphate (1302) | | Descriptor: | (3E)-4-hydroxybut-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-oxobutyl trihydrogen diphosphate, ... | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-12-02 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
8PKJ
 
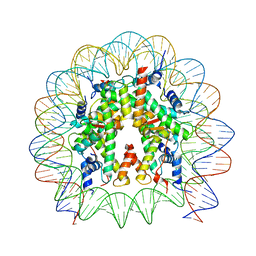 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKI
 
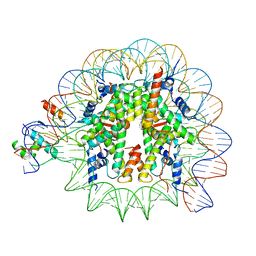 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
4PCL
 
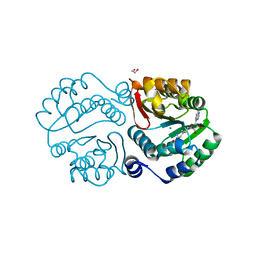 | | X-ray crystal structure of an O-methyltransferase from Anaplasma phagocytophilum bound to SAM and a Manganese ion. | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, O-methyltransferase family protein, ... | | Authors: | Fairman, J.W, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-15 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An O-Methyltransferase Is Required for Infection of Tick Cells by Anaplasma phagocytophilum.
Plos Pathog., 11, 2015
|
|
2N9C
 
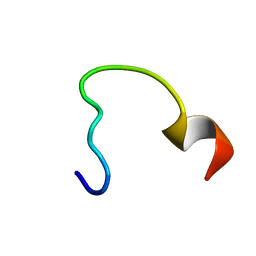 | | NRAS Isoform 5 | | Descriptor: | GTPase NRas | | Authors: | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
4PEI
 
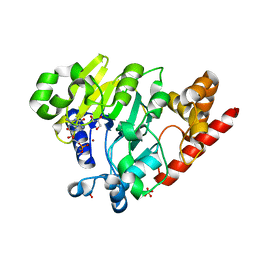 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4AGS
 
 | | Leishmania TDR1 - a unique trimeric glutathione transferase | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, THIOL-DEPENDENT REDUCTASE 1 | | Authors: | Fyfe, P.K, Westrop, G.D, Silva, A.M, Coombs, G.H, Hunter, W.N. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-04 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leishmania Tdr1 Structure, a Unique Trimeric Glutathione Transferase Capable of Deglutathionylation and Antimonial Prodrug Activation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1CIN
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(METHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
4PO5
 
 | | Crystal structure of allophycocyanin B from Synechocystis PCC 6803 | | Descriptor: | Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, PHYCOCYANOBILIN, ... | | Authors: | Pang, P.P, Dong, L.L, Sun, Y.F, Zeng, X.L, Ding, W.L, Scheer, H, Yang, X, Zhao, K.H. | | Deposit date: | 2014-02-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1OGT
 
 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE VASOACTIVE INTESTINAL PEPTIDE TYPE 1 RECEPTOR (VIPR) PEPTIDE (RESIDUES 400-408) | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2003-05-13 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dual, HLA-B27 subtype-dependent conformation of a self-peptide.
J. Exp. Med., 199, 2004
|
|
6BK7
 
 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
6KKH
 
 | | Crystal structure of the oxalate bound malyl-CoA lyase from Roseiflexus castenholzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Tang, W.R, Wang, Z.G, Zhang, C.Y, Wang, C. | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
