4J4Y
 
 | | Triple mutant GraVN | | Descriptor: | NP protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
5WZZ
 
 | | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation | | Descriptor: | Axin-1, E3 ubiquitin-protein ligase SIAH1, ZINC ION | | Authors: | Ji, L, Jiang, B, Jiang, X, Charlat, O, Chen, A, Mickanin, C, Bauer, A, Xu, W, Yan, X.-X, Cong, F. | | Deposit date: | 2017-01-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation
Genes Dev., 31, 2017
|
|
6W20
 
 | | ClpAP Disengaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W2D
 
 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
8CKB
 
 | | Asymmetric reconstruction of the crAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Cargo protein 1 gp45, Head fiber dimer protein gp29, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural atlas of the most abundant human gut virus
Nature
|
|
8CL7
 
 | | Krokinobacter eikastus rhodopsin 2 (KR2) in dark state | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Bertrand, Q, Wranik, M, Kepa, M.W, Weinert, T, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
5WUL
 
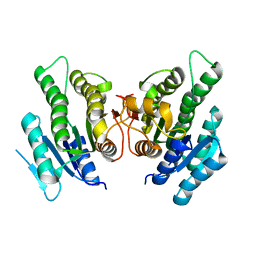 | |
3N59
 
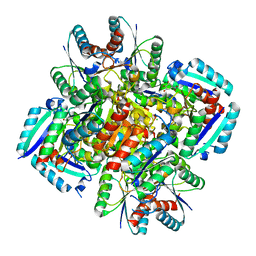 | | Type II dehydroquinase from Mycobacterium Tuberculosis complexed with 3-dehydroshikimate | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
8CL8
 
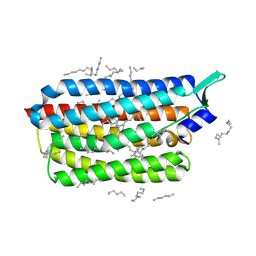 | | Krokinobacter eikastus rhodopsin 2 (KR2) extrapolated map 1us after light activation | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Bertrand, Q, Kepa, M.W, Weinert, T, Wranik, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8CL6
 
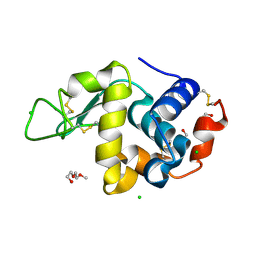 | | Lysozyme in matrix of hydroxyethylcellulose (HEC) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Bertrand, Q, Kepa, M.W, Weinert, T, Wranik, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
2OIP
 
 | |
1KSZ
 
 | | ENTRAPMENT OF 6-THIOPHOSPHORYL-IMP IN THE ACTIVE SITE OF CRYSTALLINE ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI, DATA COLLECTED AT 298K | | Descriptor: | 2-DEAZO-6-THIOPHOSPHATE GUANOSINE-5'-MONOPHOSPHATE, ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Poland, B.W, Bruns, C.A, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1997-01-14 | | Release date: | 1997-10-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Entrapment of 6-thiophosphoryl-IMP in the active site of crystalline adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 272, 1997
|
|
8CL5
 
 | | Lysozyme in matrix of lipidic cubic phase of monoolein (LCP) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Bertrand, Q, Kepa, M.W, Weinert, T, Wranik, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
6VZ8
 
 | |
3ARW
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with chelerythrine | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
5JPF
 
 | | Serine/Threonine phosphatase Z1 (Candida albicans) binds to inhibitor microcystin-LR | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Microcystin-LR, ... | | Authors: | Choy, M.S, Chen, E.H, Peti, W, Page, R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39929724 Å) | | Cite: | Molecular Insights into the Fungus-Specific Serine/Threonine Protein Phosphatase Z1 in Candida albicans.
Mbio, 7, 2016
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
4QH6
 
 | |
3ARP
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with DEQUALINIUM | | Descriptor: | Chitinase A, DEQUALINIUM, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
5SVP
 
 | | Anomalous sulfur signal reveals the position of agonist 2-methylthio-ATP bound to the ATP-gated human P2X3 ion channel in the desensitized state | | Descriptor: | 1,2-ETHANEDIOL, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5NLA
 
 | |
2OGY
 
 | | Asn199Ala Mutant of the 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase complexed with methyltetrahydrofolate to 2.3 Angstrom resolution | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION | | Authors: | Doukov, T.I, Drennan, C.L, Hemmi, H, Ragsdale, S.W. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer. Role of an active site asparagine residue in activation of methyl transfer by methyltransferases.
J.Biol.Chem., 282, 2007
|
|
5JRO
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | Descriptor: | FMN-dependent NADH-azoreductase, GLYCEROL | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-06 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
5SVK
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the ATP-bound, open state | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVT
 
 | | Anomalous Cs+ signal reveals the site of Na+ ion entry to the channel pore of the human P2X3 ion channel through the extracellular fenestrations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, P2X purinoceptor 3, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.794 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
