4R87
 
 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with CoA and spermine | | 分子名称: | COENZYME A, DI(HYDROXYETHYL)ETHER, SPERMINE, ... | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-08-29 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
8E6E
 
 | | Crystal structure of MERS 3CL protease in complex with a phenyl sulfane inhibitor | | 分子名称: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, Orf1a protein | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Madden, T.K, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
5XI5
 
 | | Crystal structure of T2R-TTL-PO5 complex | | 分子名称: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Chu, Y, Wang, Y, Yang, J, Li, W. | | 登録日 | 2017-04-26 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
To Be Published
|
|
6ZHJ
 
 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | 分子名称: | CALCIUM ION, Thermolysin, ZINC ION | | 著者 | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | 登録日 | 2020-06-23 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHN
 
 | | 3D electron diffraction structure of thaumatin from Thaumatococcus daniellii | | 分子名称: | CHLORIDE ION, Thaumatin-1 | | 著者 | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | 登録日 | 2020-06-23 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.76 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4QUW
 
 | |
4QWP
 
 | | co-crystal structure of chitosanase OU01 with substrate | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Lyu, Q, Liu, W, Han, B. | | 登録日 | 2014-07-17 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01.
Biochim.Biophys.Acta, 1850, 2015
|
|
5XS0
 
 | | Structure of a ssDNA bound to the outer DNA binding site of RAD52 | | 分子名称: | DNA repair protein RAD52 homolog, ssDNA (5'-D(*CP*CP*CP*CP*CP*C)-3'), ssDNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | 著者 | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | 登録日 | 2017-06-11 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
7ODA
 
 | | OXA-48-like Beta-lactamase OXA-436 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION | | 著者 | Lund, B.A, Thomassen, A.M, Carlsen, T.J.W, Leiros, H.K.S. | | 登録日 | 2021-04-29 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | Biochemical and biophysical characterization of the OXA-48-like carbapenemase OXA-436.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4R1Q
 
 | |
6G7B
 
 | | Nt2 domain of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | 分子名称: | ImpA-related domain protein | | 著者 | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | 登録日 | 2018-04-05 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
1SXI
 
 | | Structure of apo transcription regulator B. megaterium | | 分子名称: | Glucose-resistance amylase regulator, MAGNESIUM ION | | 著者 | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | 登録日 | 2004-03-30 | | 公開日 | 2004-10-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
6XQB
 
 | | SARS-CoV-2 RdRp/RNA complex | | 分子名称: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Liu, B, Shi, W, Yang, Y. | | 登録日 | 2020-07-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
1TLB
 
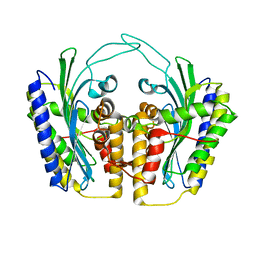 | | Yeast coproporphyrinogen oxidase | | 分子名称: | Coproporphyrinogen III oxidase, SULFATE ION | | 著者 | Phillip, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | 登録日 | 2004-06-09 | | 公開日 | 2004-07-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the oxygen-dependent coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
6FOZ
 
 | |
6XTJ
 
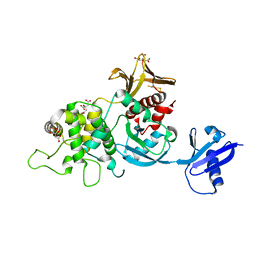 | | The high resolution structure of the FERM domain of human FERMT2 | | 分子名称: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | 著者 | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-01-16 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
6FQB
 
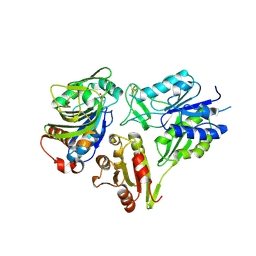 | | MurT/GatD peptidoglycan amidotransferase complex from Streptococcus pneumoniae R6 | | 分子名称: | Cobyric acid synthase, GLUTAMINE, Mur ligase family protein | | 著者 | Morlot, C, Contreras-Martel, C, Leisico, F, Straume, D, Peters, K, Hegnar, O.A, Simon, N, Villard, A.M, Breukink, E, Gravier-Pelletier, C, Le Corre, L, Vollmer, W, Pietrancosta, N, Havarstein, L.S, Zapun, A. | | 登録日 | 2018-02-13 | | 公開日 | 2018-08-22 | | 最終更新日 | 2018-11-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
6XUY
 
 | |
7NTF
 
 | |
6X5Z
 
 | | Bovine Cardiac Myosin in Complex with Chicken Skeletal Actin and Human Cardiac Tropomyosin in the Rigor State | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Doran, M.H, Lehman, W, Rynkiewicz, M.J, Bullitt, E. | | 登録日 | 2020-05-27 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM and Molecular Docking Shows Myosin Loop 4 Contacts Actin and Tropomyosin on Thin Filaments.
Biophys.J., 119, 2020
|
|
7OG2
 
 | |
1T9D
 
 | | Crystal Structure Of Yeast Acetohydroxyacid Synthase In Complex With A Sulfonylurea Herbicide, Metsulfuron methyl | | 分子名称: | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE, Acetolactate synthase, mitochondrial, ... | | 著者 | McCourt, J.A, Pang, S.S, Guddat, L.W, Duggleby, R.G. | | 登録日 | 2004-05-16 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase.
Biochemistry, 44, 2005
|
|
4PEE
 
 | | Crystal structure of a bacterial fucosidase with inhibitor 1-phenyl-4-[(2S,3S,4R,5S)-3,4-dihydroxy-5-methylpyrrolidin-2-yl]triazole | | 分子名称: | (2S,3R,4S,5S)-2-methyl-5-(1-phenyl-1H-1,2,3-triazol-4-yl)pyrrolidine-3,4-diol, Alpha-L-fucosidase, IMIDAZOLE, ... | | 著者 | Wright, D.W, Davies, G.J, Behr, J.B. | | 登録日 | 2014-04-23 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Exploiting the Hydrophobic Terrain in Fucosidases with Aryl-Substituted Pyrrolidine Iminosugars.
Chembiochem, 16, 2015
|
|
4PJX
 
 | | Structure of human MR1-Ac-6-FP in complex with human MAIT C-A11 TCR | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | 著者 | Birkinshaw, R.W, Rossjohn, J. | | 登録日 | 2014-05-12 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
6XCH
 
 | |
