1W5N
 
 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations D131C and D139C) | | 分子名称: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, ... | | 著者 | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | 登録日 | 2004-08-09 | | 公開日 | 2005-01-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
6BF6
 
 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
7WBI
 
 | | BF2*1901-FLU | | 分子名称: | Beta-2-microglobulin, ILE-ARG-HIS-GLU-ASN-ARG-MET-VAL-LEU, MHC class I alpha chain 2 | | 著者 | Liu, W.J. | | 登録日 | 2021-12-16 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7WBG
 
 | | BF2*1901/RY8 | | 分子名称: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I alpha chain 2 | | 著者 | Liu, W.J. | | 登録日 | 2021-12-16 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | 分子名称: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | 登録日 | 2010-08-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
1G0G
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152A | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
6B7Z
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | 分子名称: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2018-01-10 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5JN9
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and ethoxyzolamide | | 分子名称: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | 著者 | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | 登録日 | 2016-04-29 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
1G0K
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152C | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
3K5V
 
 | | Structure of Abl kinase in complex with imatinib and GNF-2 | | 分子名称: | 3-(6-{[4-(trifluoromethoxy)phenyl]amino}pyrimidin-4-yl)benzamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, ... | | 著者 | Cowan-Jacob, S.W, Fendrich, G, Rummel, G, Strauss, A. | | 登録日 | 2009-10-08 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors.
Nature, 463, 2010
|
|
6V3D
 
 | |
1G2W
 
 | | E177S MUTANT OF THE PYRIDOXAL-5'-PHOSPHATE ENZYME D-AMINO ACID AMINOTRANSFERASE | | 分子名称: | ACETATE ION, D-ALANINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Lepore, B.W, Ringe, D. | | 登録日 | 2000-10-21 | | 公開日 | 2000-11-15 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Studies on an Active Site Residue, E177, That Affects Binding of the Coenzyme in D-Amino Acid Transaminase, and Mechanistic Studies on a Suicide Substrate
Biochemistry and Molecular Biology of Vitamin B6 and PQQ-dependent Proteins, 10th Annual International Symposium on Vitamin B6, 2000
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
1G1V
 
 | | T4 LYSOZYME MUTANT C54T/C97A/I58T | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-13 | | 公開日 | 2001-05-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
3OFT
 
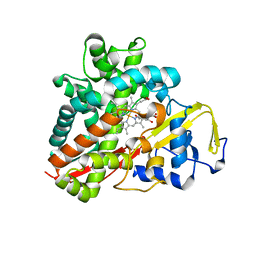 | | Crystal Structure of Cytochrome P450 CYP101C1 | | 分子名称: | (2R,5R)-hexane-2,5-diol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | 登録日 | 2010-08-16 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | 分子名称: | COLD SHOCK PROTEIN TMCSP | | 著者 | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | 登録日 | 2000-11-07 | | 公開日 | 2001-11-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
5OJR
 
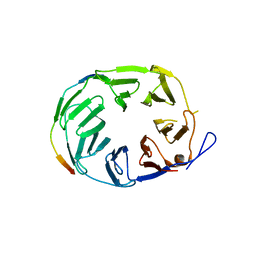 | | YCF48 bound to D1 peptide | | 分子名称: | Photosystem II protein D1 3, Ycf48-like protein | | 著者 | Michoux, F, Nixon, P.J, Murray, J.W, Bialek, W, Thieulin-Pardo, G. | | 登録日 | 2017-07-23 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1GJW
 
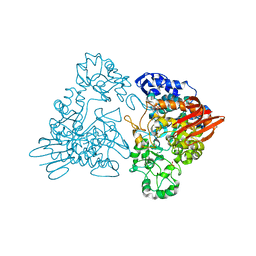 | | Thermotoga maritima maltosyltransferase complex with maltose | | 分子名称: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION, alpha-D-glucopyranose, ... | | 著者 | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | 登録日 | 2001-08-03 | | 公開日 | 2001-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
8W77
 
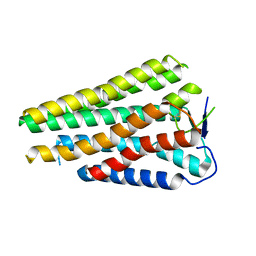 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c only | | 分子名称: | Human Consensus Olfactory Receptor OR52c in apo state, receptor only,Soluble cytochrome b562 | | 著者 | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | 登録日 | 2023-08-30 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
1FXH
 
 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | 分子名称: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | 著者 | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2000-09-26 | | 公開日 | 2001-03-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
8X23
 
 | | The Crystal Structure of MAPK13 from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Mitogen-activated protein kinase 13 | | 著者 | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | 登録日 | 2023-11-09 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Crystal Structure of MAPK13 from Biortus.
To Be Published
|
|
8X2T
 
 | |
8X2A
 
 | | The Crystal Structure of BMX from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, 4-[(3S)-3-{[(2E)-but-2-enoyl]amino}piperidin-1-yl]-5-fluoro-2,3-dimethyl-1H-indole-7-carboxamide, CHLORIDE ION, ... | | 著者 | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | 登録日 | 2023-11-09 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The Crystal Structure of BMX from Biortus.
To Be Published
|
|
1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | 分子名称: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | 著者 | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | 登録日 | 2001-08-07 | | 公開日 | 2002-01-01 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
