8UO7
 
 | |
1CCU
 
 | |
1CDG
 
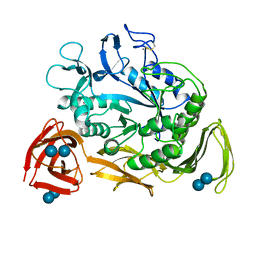 | | NUCLEOTIDE SEQUENCE AND X-RAY STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 IN A MALTOSE-DEPENDENT CRYSTAL FORM | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lawson, C.L, Van Montfort, R, Strokopytov, B.V, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1993-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form.
J.Mol.Biol., 236, 1994
|
|
3JA9
 
 | |
3JR2
 
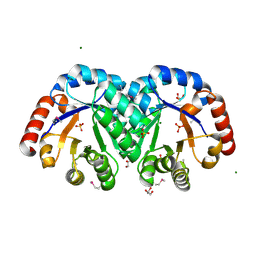 | | X-ray crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hexulose-6-phosphate synthase SgbH, ... | | Authors: | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate
decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
6BNJ
 
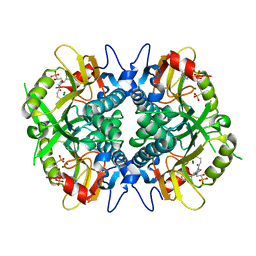 | | Human hypoxanthine guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((R)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | (3-{(3R,4R)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-[(2R)-2-hydroxy-2-phosphonoethoxy]pyrrolidin-1-yl}-3-oxopropy l)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Keough, D.T, Rejman, D, Guddat, L.W. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 13, 2018
|
|
5F1Q
 
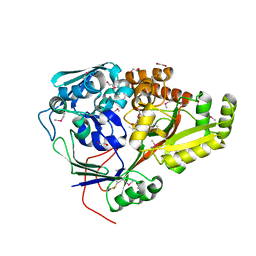 | | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis
To Be Published
|
|
3JAA
 
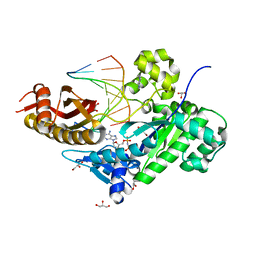 | | HUMAN DNA POLYMERASE ETA in COMPLEX WITH NORMAL DNA AND INCO NUCLEOTIDE (NRM) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3, DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Lau, W.C.Y, Li, Y, Zhang, Q, Huen, M.S.Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA
Sci Rep, 5, 2015
|
|
1C9T
 
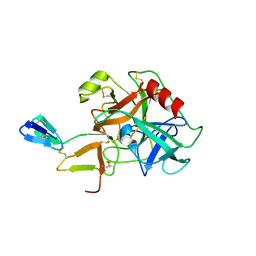 | | COMPLEX OF BDELLASTASIN WITH BOVINE TRYPSIN | | Descriptor: | BDELLASTASIN, TRYPSIN | | Authors: | Rester, U, Bode, W, Moser, M, Parry, M.A, Huber, R, Auerswald, E. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the complex of the antistasin-type inhibitor bdellastasin with trypsin and modelling of the bdellastasin-microplasmin system.
J.Mol.Biol., 293, 1999
|
|
1NPC
 
 | |
7TUU
 
 | |
1CCT
 
 | |
8U24
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 9 | | Descriptor: | Green Fluorescent Protein Variant #9, ccGFP 9 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
4BQM
 
 | | Crystal structure of human liver-type glutaminase, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTAMINASE LIVER ISOFORM, ... | | Authors: | Ferreira, I.M, Vollmar, M, Krojer, T, Strain-Damerell, C, Froese, S, Coutandin, D, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Dias, S.M.G, Ambrosio, A.L.B, Yue, W.W. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Human Liver-Type Glutaminase, Catalytic Domain
To be Published
|
|
7BGM
 
 | |
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
1NWN
 
 | |
1R4A
 
 | | Crystal Structure of GTP-bound ADP-ribosylation Factor Like Protein 1 (Arl1) and GRIP Domain of Golgin245 COMPLEX | | Descriptor: | ADP-ribosylation factor-like protein 1, Golgi autoantigen, golgin subfamily A member 4, ... | | Authors: | Wu, M, Lu, L, Hong, W, Song, H. | | Deposit date: | 2003-10-04 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4BDD
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
2VG2
 
 | | Rv2361 with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
1CB1
 
 | |
7ZUB
 
 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | Authors: | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
8A55
 
 | |
3FIX
 
 | | Crystal structure of a putative n-acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYLTRANSFERASE | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
