1R4F
 
 | | Inosine-Adenosine-Guanosine Preferring Nucleoside Hydrolase From Trypanosoma vivax: Trp260Ala Mutant In Complex With 3-Deaza-Adenosine | | 分子名称: | 3-DEAZA-ADENOSINE, CALCIUM ION, IAG-nucleoside hydrolase | | 著者 | Versees, W, Loverix, S, Vandemeulebroucke, A, Geerlings, P, Steyaert, J. | | 登録日 | 2003-10-06 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Leaving group activation by aromatic stacking: an alternative to general Acid catalysis.
J.Mol.Biol., 338, 2004
|
|
1HOZ
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX | | 分子名称: | CALCIUM ION, GLYCEROL, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | 著者 | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | 登録日 | 2000-12-12 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | 分子名称: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | 著者 | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | 登録日 | 2000-12-12 | | 公開日 | 2001-12-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1KIE
 
 | | Inosine-adenosine-guanosine preferring nucleoside hydrolase from Trypanosoma vivax: Asp10Ala mutant in complex with 3-deaza-adenosine | | 分子名称: | 3-DEAZA-ADENOSINE, CALCIUM ION, NICKEL (II) ION, ... | | 著者 | Versees, W, Decanniere, K, Van Holsbeke, E, Devroede, N, Steyaert, J. | | 登録日 | 2001-12-03 | | 公開日 | 2002-05-15 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax.
J.Biol.Chem., 277, 2002
|
|
1KIC
 
 | | Inosine-adenosine-guanosine preferring nucleoside hydrolase from Trypanosoma vivax: Asp10Ala mutant in complex with inosine | | 分子名称: | CALCIUM ION, INOSINE, NICKEL (II) ION, ... | | 著者 | Versees, W, Decanniere, K, Van Holsbeke, E, Devroede, N, Steyaert, J. | | 登録日 | 2001-12-03 | | 公開日 | 2002-05-15 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax.
J.Biol.Chem., 277, 2002
|
|
2FF1
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase soaked with ImmucillinH | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase | | 著者 | Versees, W, Barlow, J, Steyaert, J. | | 登録日 | 2005-12-18 | | 公開日 | 2006-05-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
2FF2
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase co-crystallized with ImmucillinH | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase, ... | | 著者 | Versees, W, Barlow, J, Steyaert, J. | | 登録日 | 2005-12-18 | | 公開日 | 2006-05-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
3EPX
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase in complex with the inhibitor (2R,3R,4S)-2-(hydroxymethyl)-1-(quinolin-8-ylmethyl)pyrrolidin-3,4-diol | | 分子名称: | (2R,3R,4S)-2-(hydroxymethyl)-1-(quinolin-8-ylmethyl)pyrrolidine-3,4-diol, CALCIUM ION, GLYCEROL, ... | | 著者 | Versees, W, Goeminne, A, Berg, M, Vandemeulebroucke, A, Haemers, A, Augustyns, K, Steyaert, J. | | 登録日 | 2008-09-30 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structures of T. vivax nucleoside hydrolase in complex with new potent and specific inhibitors.
Biochim.Biophys.Acta, 1794, 2009
|
|
3EPW
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase in complex with the inhibitor (2R,3R,4S)-1-[(4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-2-(hydroxymethyl)pyrrolidin-3,4-diol | | 分子名称: | 7-(((2R,3R,4S)-3,4-dihydroxy-2-(hydroxymethyl)pyrrolidin-1-yl)methyl)-3H-pyrrolo[3,2-d]pyrimidin-4(5H)-one, CALCIUM ION, IAG-nucleoside hydrolase, ... | | 著者 | Versees, W, Goeminne, A, Berg, M, Vandemeulebroucke, A, Haemers, A, Augustyns, K, Steyaert, J. | | 登録日 | 2008-09-30 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structures of T. vivax nucleoside hydrolase in complex with new potent and specific inhibitors.
Biochim.Biophys.Acta, 1794, 2009
|
|
5MJ7
 
 | | Structure of the C. elegans nucleoside hydrolase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Uncharacterized protein | | 著者 | Versees, W, Singh, R.K. | | 登録日 | 2016-11-30 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and biochemical characterization of the nucleoside hydrolase from C. elegans reveals the role of two active site cysteine residues in catalysis.
Protein Sci., 26, 2017
|
|
2NXW
 
 | | Crystal structure of phenylpyruvate decarboxylase of Azospirillum brasilense | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Versees, W, Spaepen, S, Vanderleyden, J, Steyaert, J. | | 登録日 | 2006-11-20 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. Implications for its catalytic and regulatory mechanism.
Febs J., 274, 2007
|
|
2Q5Q
 
 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP and 5-phenyl-2-oxo-valeric acid | | 分子名称: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, 5-PHENYL-2-KETO-VALERIC ACID, GLYCEROL, ... | | 著者 | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | 登録日 | 2007-06-01 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5L
 
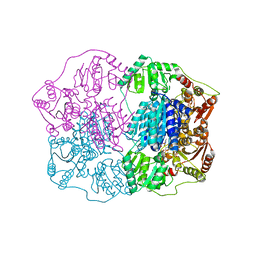 | | X-ray structure of phenylpyruvate decarboxylase in complex with 2-(1-hydroxyethyl)-3-deaza-ThDP | | 分子名称: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1S)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | 登録日 | 2007-06-01 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5J
 
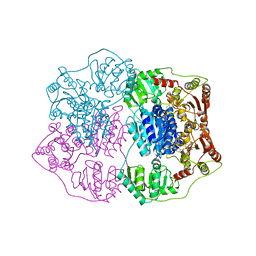 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP | | 分子名称: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Phenylpyruvate decarboxylase | | 著者 | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | 登録日 | 2007-06-01 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
2Q5O
 
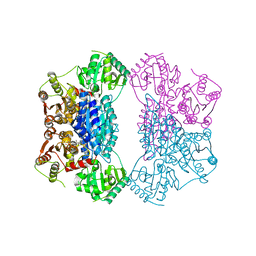 | | X-ray structure of phenylpyruvate decarboxylase in complex with 3-deaza-ThDP and phenylpyruvate | | 分子名称: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, 3-PHENYLPYRUVIC ACID, GLYCEROL, ... | | 著者 | Versees, W, Spaepen, S, Wood, M.D, Leeper, F.J, Vanderleyden, J, Steyaert, J. | | 登録日 | 2007-06-01 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase.
J.Biol.Chem., 282, 2007
|
|
6R82
 
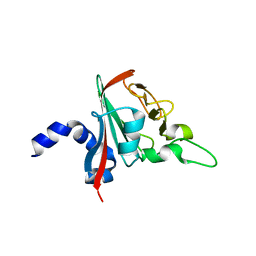 | |
3CP2
 
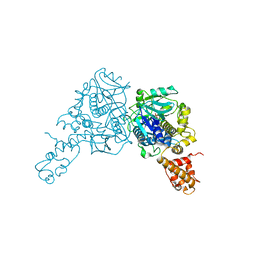 | | Crystal structure of GidA from E. coli | | 分子名称: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | 著者 | Scrima, A, Meyer, S, Versees, W, Wittinghofer, A. | | 登録日 | 2008-03-30 | | 公開日 | 2008-06-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
5HJQ
 
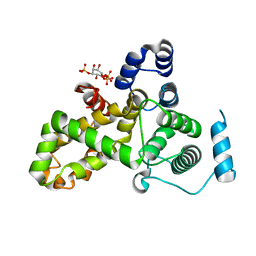 | | Crystal structure of the TBC domain of Skywalker/TBC1D24 from Drosophila melanogaster in complex with inositol(1,4,5)triphosphate | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, LD10117p | | 著者 | Fischer, B, Paesmans, J, Versees, W. | | 登録日 | 2016-01-13 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5A7T
 
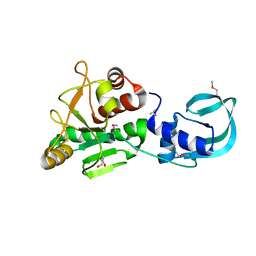 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.4 angstrom resolution. | | 分子名称: | DI(HYDROXYETHYL)ETHER, TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | 著者 | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | 登録日 | 2015-07-09 | | 公開日 | 2016-01-13 | | 最終更新日 | 2017-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5A7Z
 
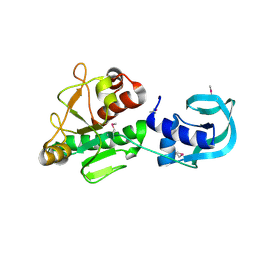 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.1 angstrom resolution. | | 分子名称: | TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | 著者 | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | 登録日 | 2015-07-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
7A0V
 
 | | Crystal structure of the 5-phosphatase domain of Synaptojanin1 in complex with a nanobody | | 分子名称: | GLYCEROL, MAGNESIUM ION, Nanobody 13015, ... | | 著者 | Paesmans, J, Galicia, C, Martin, E, Versees, W. | | 登録日 | 2020-08-11 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations.
Elife, 9, 2020
|
|
8R4C
 
 | |
8R4D
 
 | |
5HJN
 
 | |
6HLU
 
 | |
