4WKO
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hydroxybutylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-hydroxybutyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKC
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TETRAETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
2FE1
 
 | | Crystal Structure of PAE0151 from Pyrobaculum aerophilum | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bunker, R.D, Baker, E.N, Arcus, V.L. | | Deposit date: | 2005-12-15 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon.
Proteins, 72, 2008
|
|
4WKP
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 2-(2-hydroxyethoxy)ethylthiomethyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-(2-{[2-(2-hydroxyethoxy)ethyl]sulfanyl}ethyl)pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, SULFATE ION | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
2FMH
 
 | | Crystal structure of Mg2+ and BeF3- bound CheY in complex with CheZ 200-214 solved from a F432 crystal grown in Tris (pH 8.4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation.
J.Mol.Biol., 359, 2006
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
5KB0
 
 | | Crystal Structure of a Tris-thiolate Pb(II) Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5K9H
 
 | | Crystal structure of a glycoside hydrolase 29 family member from an unknown rumen bacterium | | Descriptor: | 0940_GH29, GLYCEROL, SODIUM ION, ... | | Authors: | Summers, E.L, Arcus, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8CIS
 
 | | The FERM domain of human moesin with two bound peptides identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C3P, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIR
 
 | | The FERM domain of human moesin with a bound peptide identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIU
 
 | | The FERM domain of human moesin mutant H288A | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIT
 
 | | The FERM domain of human moesin mutant L281R | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
2X6P
 
 | | Crystal Structure of Coil Ser L19C | | Descriptor: | COIL SER L19C, ZINC ION | | Authors: | Chakraborty, S, Touw, D.S, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-02-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Comparisons of Apo- and Metalated Three-Stranded Coiled Coils Clarify Metal Binding Determinants in Thiolate Containing Designed Peptides.
J.Am.Chem.Soc., 132, 2010
|
|
7L1A
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor, di-imido triphosphate (PNPNP) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, MAGNESIUM ION, ... | | Authors: | Niland, C.N, Fedorov, E, Schramm, V.L, Ghosh, A. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A.
Biochemistry, 60, 2021
|
|
2X7S
 
 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | Descriptor: | (13ALPHA,14BETA,17ALPHA)-3-HYDROXY-2-METHOXYESTRA-1,3,5(10)-TRIEN-17-YL SULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Nonsteroidal Inhibitors.
Biochemistry, 49, 2010
|
|
4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
2X7U
 
 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | Descriptor: | (9BETA,14BETA,17BETA)-17-HYDROXY-2-METHOXYESTRA-1,3,5(10)-TRIEN-3-YL SULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Non-Steroidal Inhibitors.
Biochemistry, 49, 2010
|
|
2WTN
 
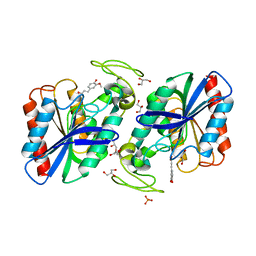 | | Ferulic Acid bound to Est1E from Butyrivibrio proteoclasticus | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, EST1E, GLYCEROL, ... | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
1I80
 
 | | CRYSTAL STRUCTURE OF M. TUBERCULOSIS PNP IN COMPLEX WITH IMINORIBITOL, 9-DEAZAHYPOXANTHINE AND PHOSPHATE ION | | Descriptor: | 9-DEAZAHYPOXANTHINE, IMINORIBITOL, PHOSPHATE ION, ... | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
7LKJ
 
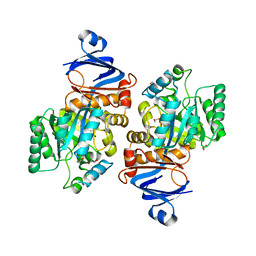 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) | | Descriptor: | 1,2-ETHANEDIOL, Aminofutalosine deaminase, FE (III) ION | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
7LKK
 
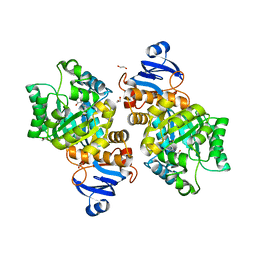 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) in complex with Methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 1,2-ETHANEDIOL, Aminofutalosine deaminase, ... | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
1K27
 
 | | Crystal Structure of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase in Complex with a Transition State Analogue | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-Deoxy-5'-Methylthioadenosine Phosphorylase, PHOSPHATE ION | | Authors: | Shi, W, Singh, V, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-09-26 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Picomolar transition state analogue inhibitors of human 5'-methylthioadenosine phosphorylase and X-ray structure with MT-immucillin-A
Biochemistry, 43, 2004
|
|
7LOU
 
 | | Crystal structure of Clostridium difficile Toxin B (TcdB) glucosyltransferase in complex with UDP and isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Glucosyltransferase TcdB, ... | | Authors: | Harijan, R.K, Paparella, A.S, Aboulache, B.L, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Inhibition of Clostridium difficile TcdA and TcdB toxins with transition state analogues.
Nat Commun, 12, 2021
|
|
7L1U
 
 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Natural Peptide-Agonist Orexin B (OxB) | | Descriptor: | Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
7L1V
 
 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Small-Molecule Agonist Compound 1 | | Descriptor: | 4'-methoxy-N,N-dimethyl-3'-{[3-(2-{[2-(2H-1,2,3-triazol-2-yl)benzene-1-carbonyl]amino}ethyl)phenyl]sulfamoyl}[1,1'-biphenyl]-3-carboxamide, Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
