2WYF
 
 | | Crystal structure of PA-IL lectin complexed with aGal12bGal-O-Met at 2.4 A resolution | | Descriptor: | CALCIUM ION, PA-I GALACTOPHILIC LECTIN, alpha-D-galactopyranose, ... | | Authors: | Nurisso, A, Blanchard, B, Varrot, A, Imberty, A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Role of Water Molecules in Structure and Energetics of Pseudomonas Aeruginosa Lectin I Interacting with Disaccharides.
J.Biol.Chem., 285, 2010
|
|
1CR3
 
 | | SOLUTION CONFORMATION OF THE (+)TRANS-ANTI-BENZO[G]CHRYSENE-DA ADDUCT OPPOSITE DT IN A DNA DUPLEX | | Descriptor: | BENZO[G]CHRYSENE, DNA (5'-D(*CP*TP*CP*TP*CP*AP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*TP*GP*AP*GP*AP*G)-3') | | Authors: | Suri, A.K, Mao, B, Amin, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1999-08-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex.
J.Mol.Biol., 292, 1999
|
|
3BWJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-(6-{[(1R)-4-carbamimidamido-1-{[(1R)-4-carbamimidamido-1-carbamoylbutyl]carbamoyl}butyl]amino}-6-oxohexyl)-3,4-dihydroxytetrahydrofuran-2-carboxamide, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Lavogina, D, Koenig, N, Uri, A, Bossemeyer, D. | | Deposit date: | 2008-01-09 | | Release date: | 2009-02-03 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases.
J.Med.Chem., 52, 2009
|
|
5IZF
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1408 | | Descriptor: | 6J9-ZEU-DAR-ACA-DAR-NH2, SULFATE ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
5IZJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1411 | | Descriptor: | 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine, 47P-AZ1-DAR-DAR, 47P-AZ1-DAR-DAR-DAR, ... | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
5J5X
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1416 | | Descriptor: | 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine, 47P-AZ1-DAL-DAR-DAR-DAR-DAR, SULFATE ION, ... | | Authors: | Alam, K.A, Ivan, T, Uri, A, Engh, R.A. | | Deposit date: | 2016-04-04 | | Release date: | 2016-07-20 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
5HTC
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5HTB
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3353 | | Descriptor: | (3R)-4-amino-3-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}-4-oxobutanoic acid (non-preferred name), (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-11 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7OOX
 
 | | Crystal structure of PIM1 in complex with ARC-3126 | | Descriptor: | 1,2-ETHANEDIOL, Inhibitor ARC-3126, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
7OOV
 
 | | Crystal structure of PIM1 in complex with ARC-1411 | | Descriptor: | 1,2-ETHANEDIOL, Inhibitor ARC-1411, SULFATE ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
7OOW
 
 | | Crystal structure of PIM1 in complex with ARC-1415 | | Descriptor: | 1,2-ETHANEDIOL, INHIBITOR ARC-1415, MAGNESIUM ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
4FBX
 
 | | Complex structure of human protein kinase CK2 catalytic subunit crystallized in the presence of a bisubstrate inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, bisubstrate inhibitor | | Authors: | Enkvist, E, Viht, K, Bischoff, N, Vahter, J, Saaver, S, Raidaru, G, Issinger, O.-G, Niefind, K, Uri, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A subnanomolar fluorescent probe for protein kinase CK2 interaction studies.
Org.Biomol.Chem., 10, 2012
|
|
3AG9
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 | | Descriptor: | (10R,20R,23R)-10-(4-aminobutyl)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamidopropyl)-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Lavogina, D, Uri, A, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2010-03-26 | | Release date: | 2010-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
3AGM
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-670 | | Descriptor: | N~2~-{8-OXO-8-[4-(9H-PURIN-6-YL)PIPERAZIN-1-YL]OCTANOYL}-D-ARGINYL-D-ARGINYL-D-ARGINYL-D-ARGINYL-D-ARGINYL-D-ARGININAMIDE, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Ragozina, J, Uri, A, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-04-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
3AGL
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1039 | | Descriptor: | (10R,20R,23R)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamido propyl)-10-methyl-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Ragozina, J, Uri, A, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-04-02 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
2IDV
 
 | | Crystal structure of wheat C113S mutant EIF4E bound TO 7-methyl-GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Dutt-Chaudhuri, A, Sadow, J, Dhaliwal, S, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
1QCR
 
 | | CRYSTAL STRUCTURE OF BOVINE MITOCHONDRIAL CYTOCHROME BC1 COMPLEX, ALPHA CARBON ATOMS ONLY | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UBIQUINOL CYTOCHROME C OXIDOREDUCTASE | | Authors: | Xia, D, Yu, C.A, Kim, H, Xia, J.Z, Kachurin, A, Zhang, L, Yu, L, Deisenhofer, J. | | Deposit date: | 1997-05-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria.
Science, 277, 1997
|
|
4JLV
 
 | | Crystal structure of the chimerical protein CapA1B1 in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, C-terminal fragment of Membrane protein CapA1, Putative uncharacterized protein capB1, ... | | Authors: | Gruszczyk, J, Olivares-Illana, V, Nourikyan, J, Fleurie, A, Bechet, E, Aumont-Nicaise, M, Gueguen-Chaignon, V, Morera, S, Grangeasse, C, Nessler, S. | | Deposit date: | 2013-03-13 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative analysis of the Tyr-kinases CapB1 and CapB2 fused to their cognate modulators CapA1 and CapA2 from Staphylococcus aureus
Plos One, 8, 2013
|
|
1VYU
 
 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYV
 
 | | beta4 subunit of Ca2+ channel | | Descriptor: | CALCIUM CHANNEL BETA-4SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYT
 
 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
3OJL
 
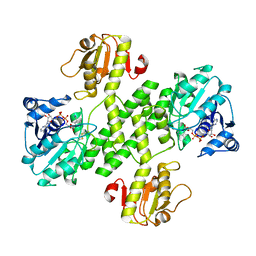 | | Native structure of the UDP-N-acetyl-mannosamine dehydrogenase Cap5O from Staphylococcus aureus | | Descriptor: | Cap5O, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nessler, S, Gruszczyk, J, Olivares-Illana, V, Meyer, P, Morera, S, Grangeasse, C, Fleurie, A. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure Analysis of the Staphylococcus aureus UDP-N-acetyl-mannosamine Dehydrogenase Cap5O Involved in Capsular Polysaccharide Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
3OGT
 
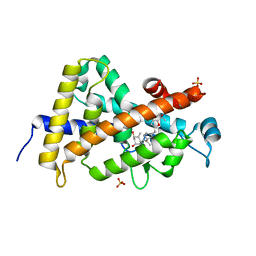 | | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3. | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Fraga, R, Mourino, A, Moras, D, Rochel, N. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3.
To be Published
|
|
2WH0
 
 | | Recognition of an intrachain tandem 14-3-3 binding site within protein kinase C epsilon | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, CALCIUM ION, PROTEIN KINASE C EPSILON TYPE, ... | | Authors: | Kostelecky, B, Saurin, A.T, Purkiss, A, Parker, P.J, McDonald, N.Q. | | Deposit date: | 2009-04-28 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Recognition of an Intra-Chain Tandem 14-3-3 Binding Site within Pkc Epsilon.
Embo Rep., 10, 2009
|
|
3CS6
 
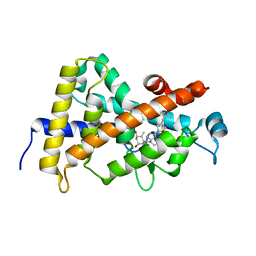 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, P, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
