8CDA
 
 | | Crystal structure of MAB_4123 from Mycobacterium abscessus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Probable monooxygenase | | Authors: | Ung, K.L, Poussineau, C, Couston, J, Alsarraf, H.M.A.B, Blaise, M. | | Deposit date: | 2023-01-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MAB_4123, a putative flavin-dependent monooxygenase from Mycobacterium abscessus.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7QPA
 
 | | Outward-facing auxin bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
7QP9
 
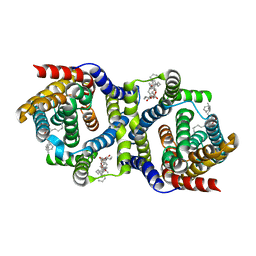 | | Outward-facing apo-form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
7QPC
 
 | | Inward-facing NPA bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
4FGL
 
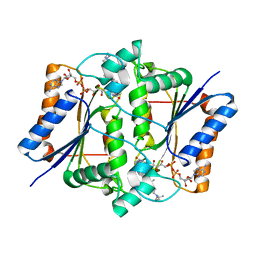 | | Reduced quinone reductase 2 in complex with chloroquine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2012-06-04 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of quinone reductase 2 bound to antimalarial drugs reveal conformational change upon reduction
J.Biol.Chem., 2013
|
|
4FGJ
 
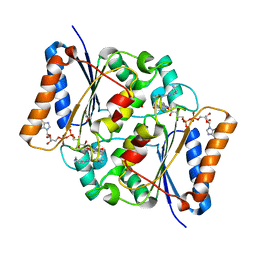 | | Oxidized quinone reductase 2 in complex with primaquine | | Descriptor: | (4S)-N~4~-(6-methoxyquinolin-8-yl)pentane-1,4-diamine, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2012-06-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Crystal structures of quinone reductase 2 bound to antimalarial drugs reveal conformational change upon reduction
J.Biol.Chem., 2013
|
|
4FGK
 
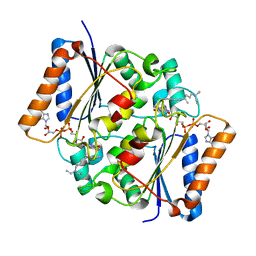 | | Oxidized quinone reductase 2 in complex with chloroquine | | Descriptor: | (4S)-N~4~-(7-chloroquinolin-4-yl)-N~1~,N~1~-diethylpentane-1,4-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2012-06-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structures of quinone reductase 2 bound to antimalarial drugs reveal conformational change upon reduction
J.Biol.Chem., 2013
|
|
5HGG
 
 | | Crystal structure of uPA in complex with a camelid-derived antibody fragment | | Descriptor: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Camelid Derived Antibody Fragment, ... | | Authors: | Yung, K.W.Y, Kromann-Hansen, T, Andreasen, P.A, Ngo, J.C.K. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-01 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
J.Biol.Chem., 291, 2016
|
|
4U7G
 
 | | Oxidized quinone reductase 2 in complex with CK2 inhibitor TBBz | | Descriptor: | 4,5,6,7-TETRABROMO-BENZIMIDAZOLE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
4U7F
 
 | | Reduced quinone reductase 2 in complex with CK2 inhibitor DMAT | | Descriptor: | 4,5,6,7-TETRABROMO-N,N-DIMETHYL-1H-BENZIMIDAZOL-2-AMINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
4U7H
 
 | | Oxidized quinone reductase 2 in complex with CK2 inhibitor DMAT | | Descriptor: | 4,5,6,7-TETRABROMO-N,N-DIMETHYL-1H-BENZIMIDAZOL-2-AMINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
7QO5
 
 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
6ITU
 
 | | Crystal Structure of the GULP1 PTB domain-APP peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 protein, PTB domain-containing engulfment adapter protein 1, ... | | Authors: | Yung, K.W.Y, Lau, K.F, Ngo, J.C.K. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Attenuation of amyloid-beta generation by atypical protein kinase C-mediated phosphorylation of engulfment adaptor PTB domain containing 1 threonine 35.
Faseb J., 33, 2019
|
|
7QO3
 
 | | Structure of the 26S proteasome-Ubp6 complex in the si state (Core Particle and Lid) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO6
 
 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
8R8Q
 
 | | Lysosomal peptide transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosylated lysosomal membrane protein, Major facilitator superfamily domain-containing protein 1 | | Authors: | Jungnickel, K.E.J, Loew, C. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes.
Nat.Cell Biol., 26, 2024
|
|
7QO4
 
 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
8Q43
 
 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-C DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*CP*CP*CP*CP*C)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q44
 
 | | Crystal structure of cA4-bound Can2 (E364R) in complex with oligo-T DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*T)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q40
 
 | | Crystal structure of cA4 activated Can2 in complex with a cleaved DNA substrate | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*CP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q41
 
 | | Crystal structure of Can2 (E341A) bound to cA4 and TTTAAA ssDNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
7UIM
 
 | |
7UIN
 
 | | CryoEM Structure of an Group II Intron Retroelement | | Descriptor: | AMMONIUM ION, DNA (37-MER), E.r IIC Intron, ... | | Authors: | Chung, K, Xu, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a mobile intron retroelement poised to attack its structured DNA target
Science, 378, 2022
|
|
6EV1
 
 | | Crystal structure of antibody against schizophyllan | | Descriptor: | Heavy chain, Light chain | | Authors: | Sung, K.H, Josewski, J, Dubel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6JF6
 
 | |
