4V15
 
 | | Crystal structure of D-threonine aldolase from Alcaligenes xylosoxidans | | Descriptor: | D-THREONINE ALDOLASE, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Uhl, M.K, Oberdorfer, G, Steinkellner, G, Riegler, L, Schuermann, M, Gruber, K. | | Deposit date: | 2014-09-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of D-Threonine Aldolase from Alcaligenes Xylosoxidans Provides Insight Into a Metal Ion Assisted Plp-Dependent Mechanism.
Plos One, 10, 2015
|
|
3GR7
 
 | |
3GR8
 
 | | Structure of OYE from Geobacillus kaustophilus, orthorhombic crystal form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, ... | | Authors: | Uhl, M.K, Gruber, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Old Yellow Enzyme-Catalyzed Dehydrogenation of Saturated Ketones
ADV.SYNTH.CATAL., 353, 2011
|
|
6FRI
 
 | |
4D7K
 
 | |
6YJ0
 
 | |
1NCT
 
 | | TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
1V1C
 
 | |
1NCU
 
 | | Titin Module M5, N-terminally Extended, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
1TNN
 
 | |
1TNM
 
 | |
1GXE
 
 | | Central domain of cardiac myosin binding protein C | | Descriptor: | MYOSIN BINDING PROTEIN C, CARDIAC-TYPE | | Authors: | Pfuhl, M. | | Deposit date: | 2002-04-03 | | Release date: | 2003-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Stability and Dynamics of the Central Domain of Cardiac Myosin Binding Protein C (Mybp-C): Implications for Multidomain Assembly and Causes for Cardiomyopathy
J.Mol.Biol., 329, 2003
|
|
5A4K
 
 | | Crystal structure of the R139W variant of human NAD(P)H:quinone oxidoreductase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Strandback, E, Gudipati, V, Uhl, M.K, Rantase, D.M, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Catalytic competence, structure and stability of the cancer-associated R139W variant of the human NAD(P)H:quinone oxidoreductase 1 (NQO1).
FEBS J., 284, 2017
|
|
4CET
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with dicoumarol at 2.2 A resolution | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
4CF6
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with Cibacron blue at 2.7 A resolution | | Descriptor: | CIBACRON BLUE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
4D1Y
 
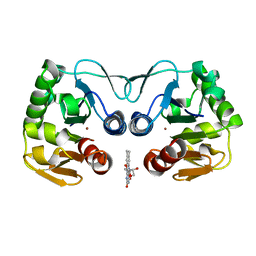 | | Crystal structure of a putative protease from Bacteroides thetaiotaomicron. | | Descriptor: | PUTATIVE PROTEASE I, RIBOFLAVIN, ZINC ION | | Authors: | Knaus, T, Uhl, M.K, Monschein, S, Moratti, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Stability of an Unusual Zinc-Binding Protein from Bacteroides Thetaiotaomicron.
Biochim.Biophys.Acta, 1844, 2014
|
|
3QUL
 
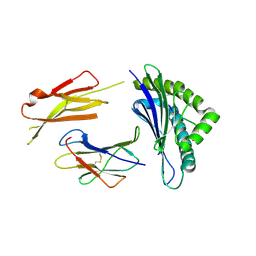 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
3QUK
 
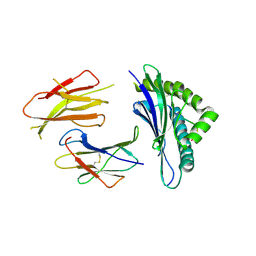 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4A) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
2UZR
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
2UZS
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
2AVG
 
 | |
1K5N
 
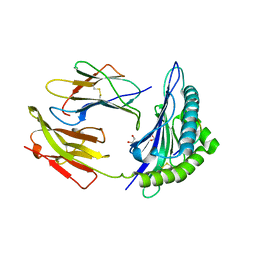 | | HLA-B*2709 BOUND TO NONA-PEPTIDE M9 | | Descriptor: | GLYCEROL, beta-2-microglobulin, light chain, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | HLA-B27 Subtypes Differentially Associated with Disease Exhibit Subtle Structural Alterations
J.Biol.Chem., 277, 2002
|
|
2K1M
 
 | |
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
