8XEU
 
 | |
8XFA
 
 | | Structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, FE (III) ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XFI
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound (crystal form 2) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, CHLORIDE ION, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XET
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF4
 
 | | Structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with Bisucaberin bound | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Iron-hydroxamate ABC transporter substrate-binding protein FtsB | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF8
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine B | | Descriptor: | 1,2-ETHANEDIOL, Ferrioxamine B, Iron-hydroxamate ABC transporter substrate-binding protein FtsB, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF9
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
1WT5
 
 | | The Crystal Structure Of A Humanized Antibody Fv 528 | | Descriptor: | ANTI EGFR ANTIBODY FV REGION | | Authors: | Makabe, K, Tsumoto, K, Asano, R, Kondo, H, Kumagai, I. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic consequences of mutations in vernier zone residues of a humanized anti-human epidermal growth factor receptor murine antibody, 528
J.Biol.Chem., 283, 2008
|
|
8JOW
 
 | |
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5CIL
 
 | | Crystal Structure of non-neutralizing version of 4E10 (WDWD) with epitope bound | | Descriptor: | ACETATE ION, CHLORIDE ION, FAB 4E10 HEAVY CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5CIP
 
 | | Crystal Structure of Unbound 4E10 | | Descriptor: | FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5CIN
 
 | | Crystal Structure of non-neutralizing version of 4E10 (DeltaLoop) with epitope bound | | Descriptor: | CHLORIDE ION, FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
8H63
 
 | | Crystal structure of Internalin A from Listeria monocytogenes with nanobody VHH10 bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Nagatoish, S, Tsumoto, K. | | Deposit date: | 2022-10-15 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
J.Biol.Chem., 299, 2023
|
|
8H64
 
 | |
8H62
 
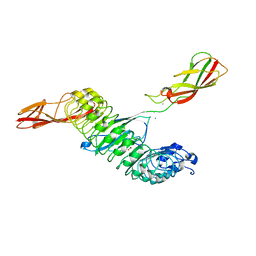 | | Crystal structure of Internalin A from Listeria monocytogenes with human E-cadherin EC12 | | Descriptor: | ACETATE ION, CALCIUM ION, Cadherin-1, ... | | Authors: | Caaveiro, J.M.M, Nagatoish, S, Tsumoto, K. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
J.Biol.Chem., 299, 2023
|
|
8HYI
 
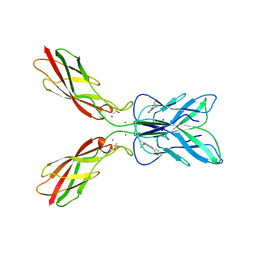 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine | | Descriptor: | 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Modulation of a conformational ensemble by a small molecule that inhibits key protein-protein interactions involved in cell adhesion.
Protein Sci., 32, 2023
|
|
1UFU
 
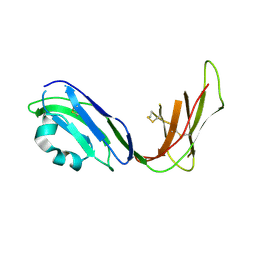 | | Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) | | Descriptor: | Immunoglobulin-like transcript 2 | | Authors: | Shiroishi, M, Amano, K, Rasubala, L, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and thermodynamic properties of the interaction between Immunoglobulin like transcript (ILT) and MHC class I
To be Published
|
|
1IC4
 
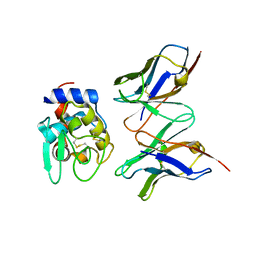 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC7
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
4P35
 
 | | Crystal structure of DJ-1 with Zinc(II) bound (crystal I) | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DJ-1, ... | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
1IC5
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
4P34
 
 | | Crystal structure of DJ-1 in sulfenic acid form (fresh crystal) | | Descriptor: | PENTAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
4P2G
 
 | | Crystal structure of DJ-1 in sulfinic acid form (aged crystal) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
4P36
 
 | | Crystal structure of DJ-1 With Zn(II) bound (crystal 2) | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Protein DJ-1, ZINC ION | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
