1WA2
 
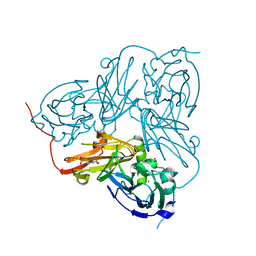 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase with nitrite bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE,, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
3KB6
 
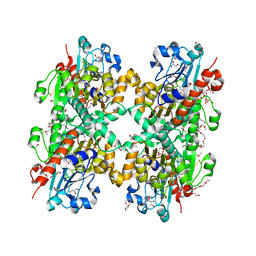 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4D6U
 
 | | Cytochrome bc1 bound to the 4(1H)-pyridone GSK932121 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, CARDIOLIPIN, ... | | Authors: | Capper, M.J, ONeill, P.M, Fisher, N, Strange, R.W, Moss, D, Ward, S.A, Berry, N.G, Lawrenson, A.S, Hasnain, S.S, Biagini, G.A, Antonyuk, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.09 Å) | | Cite: | Antimalarial 4(1H)-Pyridones Bind to the Qi Site of Cytochrome Bc1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3IXQ
 
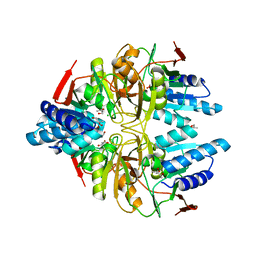 | | Structure of ribose 5-phosphate isomerase a from methanocaldococcus jannaschii | | Descriptor: | ACETATE ION, CHLORIDE ION, Ribose-5-phosphate isomerase A, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of an archaeal ribose-5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IWT
 
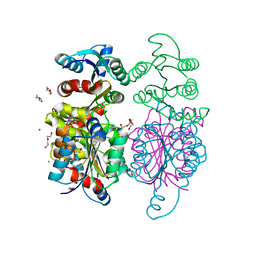 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1OZU
 
 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
4D6T
 
 | | Cytochrome bc1 bound to the 4(1H)-pyridone GW844520 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-chloro-2,6-dimethyl-5-{4-[4-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, CARDIOLIPIN, ... | | Authors: | Capper, M.J, O'Neill, P.M, Fisher, N, Strange, R.W, Moss, D, Ward, S.A, Berry, N.G, Lawrenson, A.S, Hasnain, S.S, Biagini, G.A, Antonyuk, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Antimalarial 4(1H)-Pyridones Bind to the Qi Site of Cytochrome Bc1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4D4N
 
 | | Nitrosyl complex of the D121A variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
4D4X
 
 | | Nitrosyl complex of the D121I variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-09 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
2XLM
 
 | | Cytochrome c prime from Alcaligenes xylosoxidans: Ferrous recombinant native with bound NO | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R, Hasnain, S.S. | | Deposit date: | 2010-07-21 | | Release date: | 2010-11-10 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Distal-to-Proximal No Conversion in Hemoproteins: The Role of the Proximal Pocket.
J.Mol.Biol., 405, 2011
|
|
3CQQ
 
 | | Human SOD1 G85R Variant, Structure II | | Descriptor: | ACETYL GROUP, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Cao, X, Antonyuk, S, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
1OBF
 
 | | The crystal structure of Glyceraldehyde 3-phosphate Dehydrogenase from Alcaligenes xylosoxidans at 1.7A resolution. | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Eady, R.R, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Glyceraldehyde 3-Phosphate Dehydrogenase from Alcaligenes Xylosoxidans at 1.7 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5CXZ
 
 | | SYK catalytic domain complexed with naphthyridine inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{7-[4-(dimethylamino)phenyl]-1,6-naphthyridin-5-yl}propane-1,3-diamine, ... | | Authors: | Thoma, G, Veenstra, S, Strang, R, Blanz, J, Vangrevelinghe, E, Berghausen, J, Lee, C.C, Zerwes, H.-G. | | Deposit date: | 2015-07-29 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Orally bioavailable Syk inhibitors with activity in a rat PK/PD model.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2YB9
 
 | | Crystal Structure of Human Neutral Endopeptidase complexed with a heteroarylalanine diacid. | | Descriptor: | HETEROARYLALANINE 5-PHENYL OXAZOLE, NEPRILYSIN, ZINC ION | | Authors: | Glossop, M.S, Bazin, R.J, Dack, K.N, Done, S, Fox, D.N.A, MacDonald, G.A, Mills, M, Owen, D.R, Phillips, C, Reeves, K.A, Ringer, T.J, Strang, R.S, Watson, C.A.L. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-25 | | Last modified: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Evaluation of Heteroarylalanine Diacids as Potent and Selective Neutral Endopeptidase Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2BP0
 
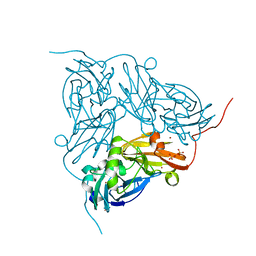 | | M144L mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-17 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2BWI
 
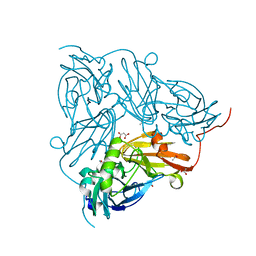 | | Atomic Resolution Structure of Nitrite -soaked Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BWD
 
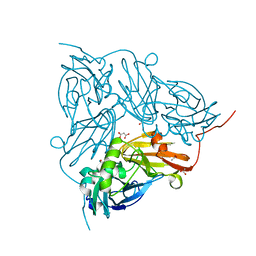 | | Atomic Resolution Structure of Achromobacter cycloclastes Cu Nitrite Reductase with Endogenously bound Nitrite and NO | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1HAU
 
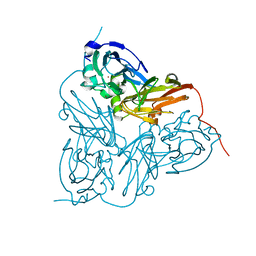 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HAW
 
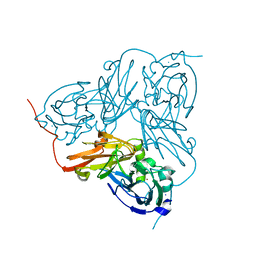 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (I) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2BW4
 
 | | Atomic Resolution Structure of Resting State of the Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BW5
 
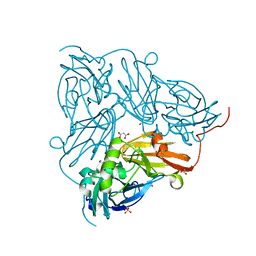 | | Atomic Resolution Structure of NO-bound Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BP8
 
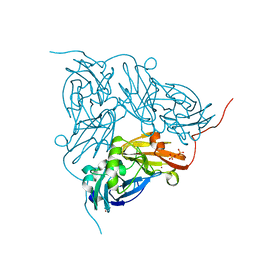 | | M144Q Structure of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-18 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
3CQP
 
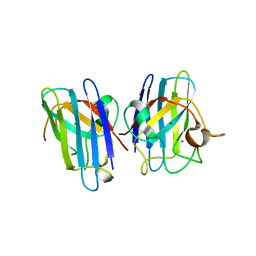 | | Human SOD1 G85R Variant, Structure I | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2JLP
 
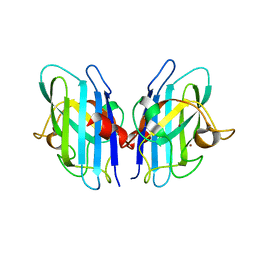 | | Crystal structure of human extracellular copper-zinc superoxide dismutase. | | Descriptor: | COPPER (II) ION, EXTRACELLULAR SUPEROXIDE DISMUTASE (CU-ZN), THIOCYANATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Marklund, S.L, Hasnain, S.S. | | Deposit date: | 2008-09-14 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Human Extracellular Copper-Zinc Superoxide Dismutase at 1.7 A Resolution: Insights Into Heparin and Collagen Binding.
J.Mol.Biol., 388, 2009
|
|
2PCR
 
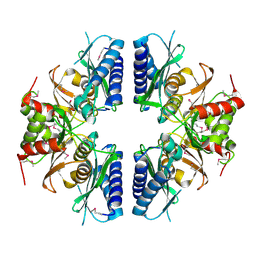 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
