1U3D
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4PK7
 
 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3U65
 
 | | The Crystal Structure of Tat-P(T) (Tp0957) | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, Tp33 protein | | Authors: | Brautigam, C.A, Tomchick, D.R, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
1U3C
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2GTD
 
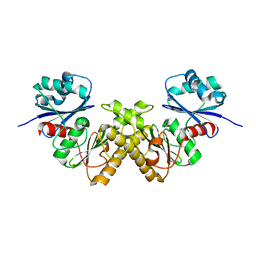 | | Crystal Structure of a Type III Pantothenate Kinase: Insight into the Catalysis of an Essential Coenzyme A Biosynthetic Enzyme Universally Distributed in Bacteria | | Descriptor: | Type III Pantothenate Kinase | | Authors: | Yang, K, Eyobo, Y, Brand, A.L, Martynowski, D, Tomchick, D. | | Deposit date: | 2006-04-27 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Type III Pantothenate Kinase: Insight into the Mechanism of an Essential Coenzyme A Biosynthetic Enzyme Universally Distributed in Bacteria.
J.Bacteriol., 188, 2006
|
|
1X80
 
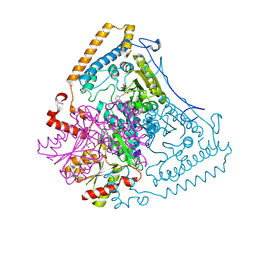 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
7KH2
 
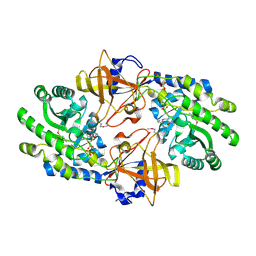 | | Structure of N-citrylornithine decarboxylase bound with PLP | | Descriptor: | GLYCEROL, N-citrylornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M, Michael, A. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-16 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Alternative pathways utilize or circumvent putrescine for biosynthesis of putrescine-containing rhizoferrin.
J.Biol.Chem., 296, 2020
|
|
1X7W
 
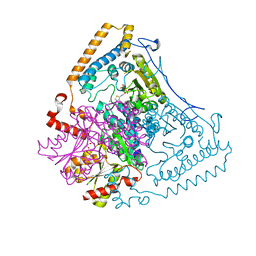 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1JPZ
 
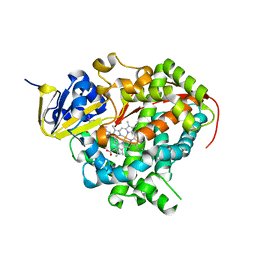 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
2OIV
 
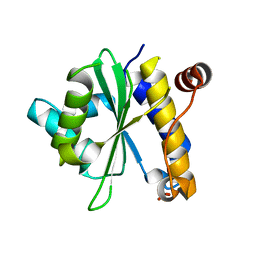 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
1X7X
 
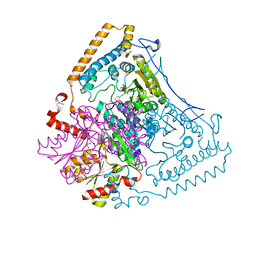 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
7L0K
 
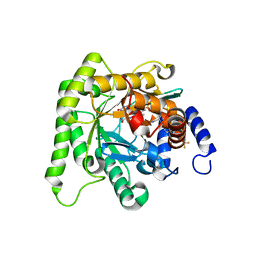 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM784 (3-(1-(3-methyl-4-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrrole-2-carboxamido)ethyl)-1H-pyrazole-5-carboxamide) | | Descriptor: | 3-{(1R)-1-[(3-methyl-4-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-1H-pyrrole-2-carbonyl)amino]ethyl}-1H-pyrazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZ4
 
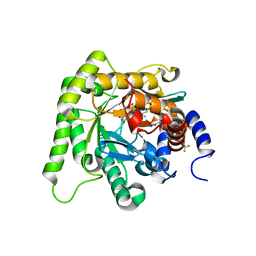 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM705 (N-(1-(1H-1,2,4-triazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(1H-1,2,4-triazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYV
 
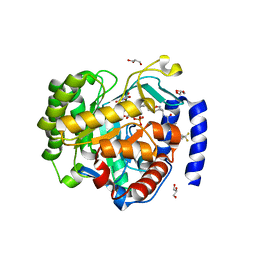 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM634 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(4-(trifluoromethyl)benzyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-{[4-(trifluoromethyl)phenyl]methyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZY
 
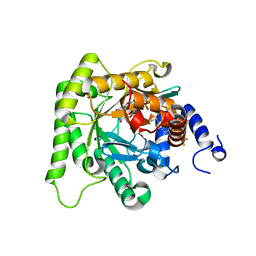 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM778 (3-methyl-N-(1-(5-methyl-1H-pyrazol-3-yl)ethyl)-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1H-pyrazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7L01
 
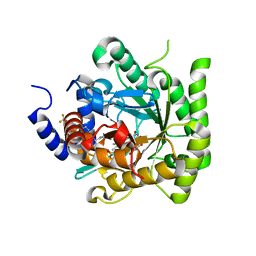 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM782 (N-(1-(5-cyano-1H-pyrazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
1KQN
 
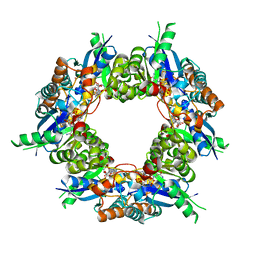 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
1KR2
 
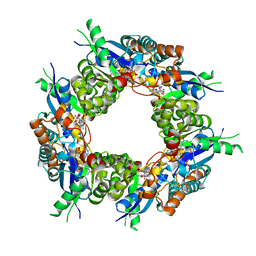 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1KQO
 
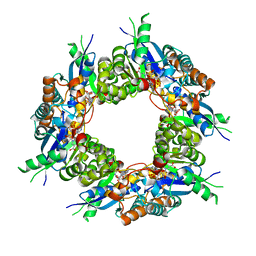 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1R61
 
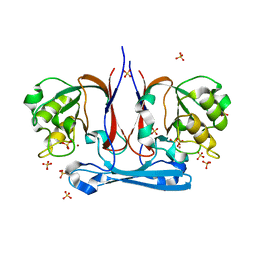 | | The structure of predicted metal-dependent hydrolase from Bacillus stearothermophilus | | Descriptor: | SULFATE ION, ZINC ION, metal-dependent hydrolase | | Authors: | Maderova, J, Borek, D, Tomchick, D, Joachimiak, A, Collart, F, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of potential metal-dependent hydrolase with cyclase activity
To be Published
|
|
1X7Z
 
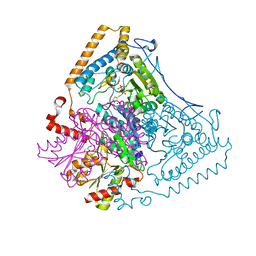 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7Y
 
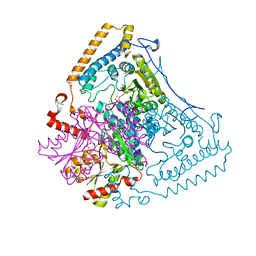 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
4PJU
 
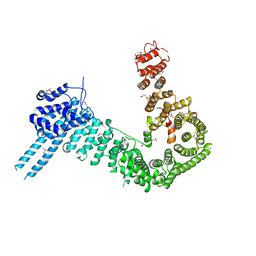 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PJW
 
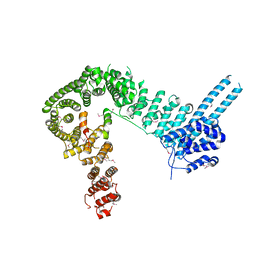 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1), with bound MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7KYK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM589 (ethyl 3-methyl-4-((4-(trifluoromethyl)benzo[d]oxazol-7-yl)methyl)-1H-pyrrole-2-carboxylate) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
