4AD0
 
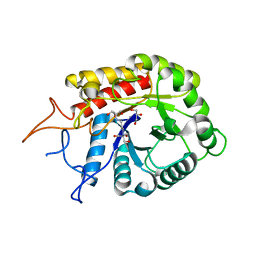 | | Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
226D
 
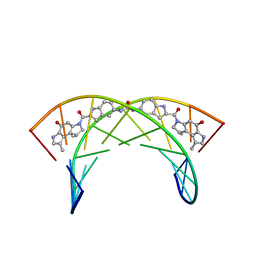 | |
1RAJ
 
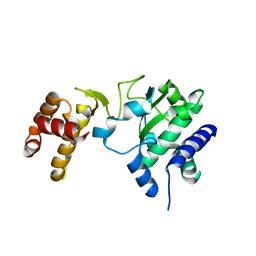 | |
1RA7
 
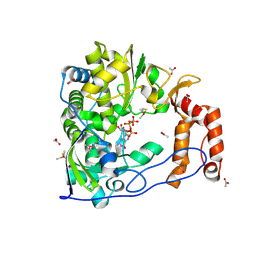 | | Poliovirus Polymerase with GTP | | Descriptor: | ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | Authors: | Thompson, A.A, Peersen, O.B. | | Deposit date: | 2003-10-31 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase.
Embo J., 23, 2004
|
|
3K3Q
 
 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
4V1S
 
 | | Structure of the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron | | Descriptor: | ALPHA-1,6-MANNANASE, GLYCEROL | | Authors: | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZJ6
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with cyclohexylmethyl-Glc-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Thompson, A.J, Sobala, L.F, Fernandes, P.Z, Hakki, Z, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5JOV
 
 | | Bacteroides ovatus Xyloglucan PUL GH31 with bound 5FIdoF | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-fluoro-alpha-L-idopyranose, ... | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JOY
 
 | | Bacteroides ovatus Xyloglucan PUL GH43A in complex with AraLOG | | Descriptor: | (Z)-L-Arabinonhydroximo-1,4-lactone, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JOX
 
 | | Bacteroides ovatus Xyloglucan PUL GH43A in complex with AraDNJ | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JOW
 
 | | Bacteroides ovatus Xyloglucan PUL GH43A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JOU
 
 | | Bacteroides ovatus Xyloglucan PUL GH31 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase BoGH31A, NICKEL (II) ION | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
6CZD
 
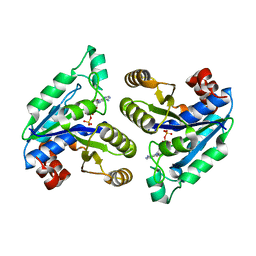 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8VXF
 
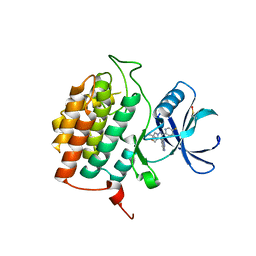 | | Structure of Casein kinase I isoform delta (CK1d) complexed with inhibitor 15 | | Descriptor: | (2P,3P,8S)-2-(5-fluoropyridin-2-yl)-6,6-dimethyl-3-(1H-pyrazolo[3,4-b]pyridin-4-yl)-6,7-dihydro-4H-pyrazolo[5,1-c][1,4]oxazine, Casein kinase I isoform delta | | Authors: | Thompson, A.A, Milligan, C.M, Sharma, S. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8VXD
 
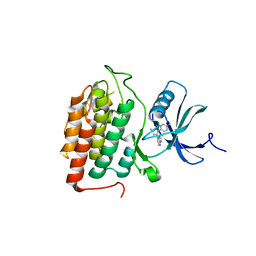 | |
6NVD
 
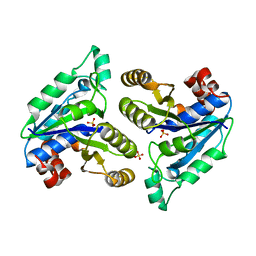 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9 | | Descriptor: | 2',5'-dideoxy-5'-[4-({[(1S,2R)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetyl}amino)-1H-1,2,3-triazol-1-yl]cytidine, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9
To Be Published
|
|
3GB7
 
 | | Potassium Channel KcsA-Fab complex in Li+ | | Descriptor: | DIACYL GLYCEROL, NICKEL (II) ION, Voltage-gated potassium channel, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-02-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6NWG
 
 | |
6NVC
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 6 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, {(1S,2R)-2-[4-(carboxymethyl)benzene-1-carbonyl]cyclopentyl}acetic acid | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 6
To Be Published
|
|
3IGA
 
 | | Potassium Channel KcsA-Fab complex in Li+ and K+ | | Descriptor: | Antibody Fab Fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8EA8
 
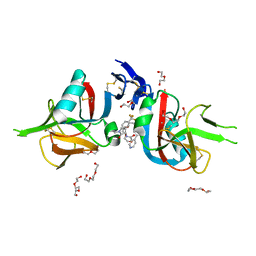 | | NKG2D complexed with inhibitor 4a | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA7
 
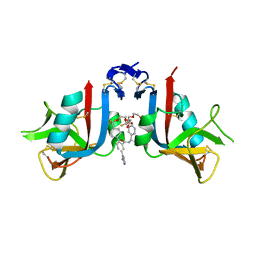 | | NKG2D complexed with inhibitor 3g | | Descriptor: | (4M)-N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-(1-methyl-1H-pyrazol-5-yl)-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, DI(HYDROXYETHYL)ETHER, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA5
 
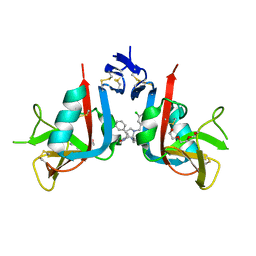 | | NKG2D complexed with inhibitor 1a | | Descriptor: | (3S,5aS,8aR)-3-benzyl-6-[(3,5-dichlorophenyl)methyl]-1,4-dimethyloctahydropyrrolo[3,2-e][1,4]diazepine-2,5-dione, NKG2-D type II integral membrane protein, TRIETHYLENE GLYCOL | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA9
 
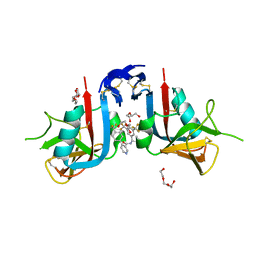 | | NKG2D complexed with inhibitor 4d | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-1-[3,5-bis(trifluoromethyl)phenyl]-2-(dimethylamino)-2-oxoethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAB
 
 | | NKG2D complexed with inhibitor 4f | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-2-oxo-1-[3-(trifluoromethyl)phenyl]-2-({4-[4-(trifluoromethyl)phenyl]pyridin-3-yl}amino)ethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
