6VGT
 
 | |
2X6G
 
 | |
5COR
 
 | |
6VE9
 
 | |
6VHJ
 
 | |
5COY
 
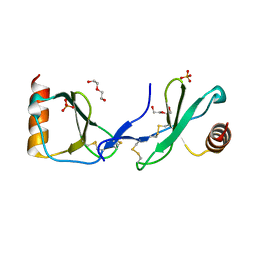 | | Crystal structure of CC chemokine 5 (CCL5) | | Descriptor: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-07-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6XOW
 
 | |
2X6L
 
 | |
2X69
 
 | |
6XOS
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOT
 
 | |
6XLY
 
 | |
1I5U
 
 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
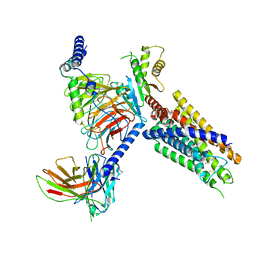 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
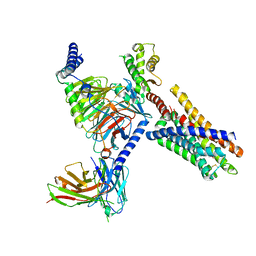 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J21
 
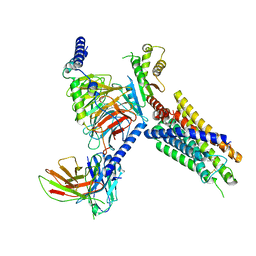 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
7K1D
 
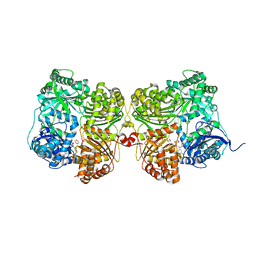 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_77291 | | Descriptor: | (3R)-3-{4-[(3R)-4-(3,4-difluorobenzene-1-carbonyl)morpholin-3-yl]-1H-1,2,3-triazol-1-yl}-N-hydroxy-4-(naphthalen-2-yl)butanamide, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 2
To Be Published
|
|
7K1F
 
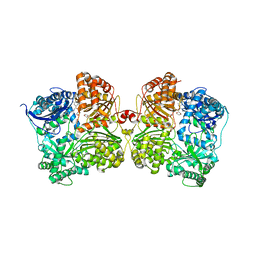 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88558 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-({1-[(2R)-4-(hydroxyamino)-4-oxo-1-(quinolin-7-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 4
To Be Published
|
|
7K1E
 
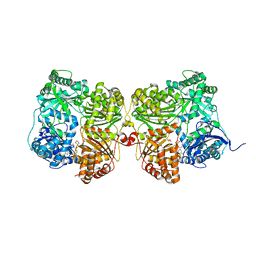 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88646 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-[(1S)-1-{1-[(2R)-4-(hydroxyamino)-4-oxo-1-(5,6,7,8-tetrahydronaphthalen-2-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}ethyl]benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 3
To Be Published
|
|
1I5T
 
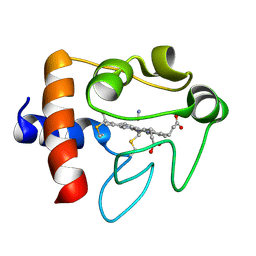 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
1M60
 
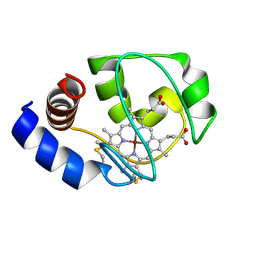 | | Solution Structure of Zinc-substituted cytochrome c | | Descriptor: | ZINC SUBSTITUTED HEME C, Zinc-substituted cytochrome c | | Authors: | Qian, C, Yao, Y, Tong, Y, Wang, J, Tang, W. | | Deposit date: | 2002-07-11 | | Release date: | 2002-08-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of zinc-substituted cytochrome c.
J.Biol.Inorg.Chem., 8, 2003
|
|
7K1V
 
 | |
