6J0I
 
 | | Structure of [Co2+-(Chromomycin A3)2]-d(TTGGCGAA)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Satange, R.B, Chuang, C.Y, Hou, M.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation.
Nucleic Acids Res., 47, 2019
|
|
6J0H
 
 | | Crystal structure of Actinomycin D- d(TTGGCGAA) complex | | Descriptor: | Actinomycin D, DNA (5'-D(P*TP*TP*GP*GP*CP*GP*AP*A)-3'), SODIUM ION | | Authors: | Satange, R.B, Hou, M.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation.
Nucleic Acids Res., 47, 2019
|
|
5GUN
 
 | | Crystal structure of d(GTGGAATGGAAC) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*TP*GP*GP*AP*AP*TP*GP*GP*AP*AP*C)-3') | | Authors: | Satange, R.B, Kao, Y.F, Hou, M.H. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2GIS
 
 | |
3IQR
 
 | |
3IQN
 
 | |
3IQP
 
 | |
3GX2
 
 | |
3GX7
 
 | |
3GX3
 
 | |
3GX5
 
 | |
3GX6
 
 | |
7DQ8
 
 | |
7DQ0
 
 | |
4H86
 
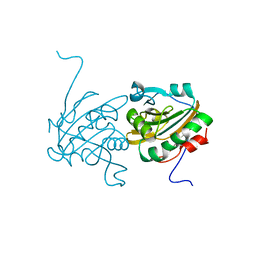 | | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Liu, M, Wang, F, Qiu, R, Wu, T, Gu, S, Tang, R, Ji, C. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form
To be Published
|
|
6JEA
 
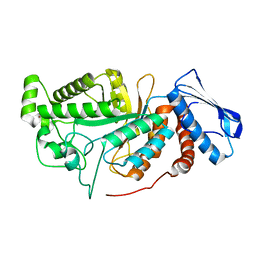 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6JE8
 
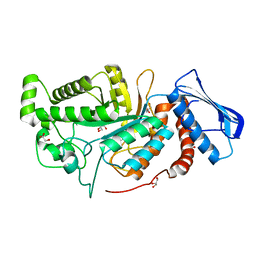 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | Beta-N-acetylhexosaminidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6JEB
 
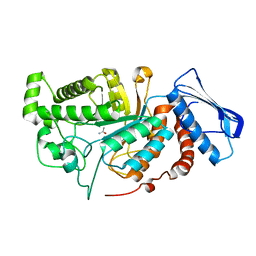 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | ACETAMIDE, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7BPV
 
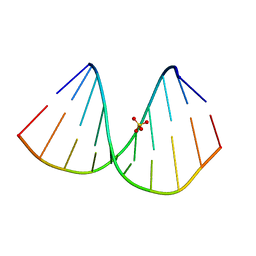 | | r(GUGGGCCGAC)/d(GTCGGCCCAC) hybrid duplex structure | | Descriptor: | DNA (5'-D(P*GP*TP*CP*GP*GP*CP*CP*CP*AP*C)-3'), RNA (5'-R(P*GP*UP*GP*GP*GP*CP*CP*GP*AP*C)-3'), SULFATE ION | | Authors: | Wang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6L76
 
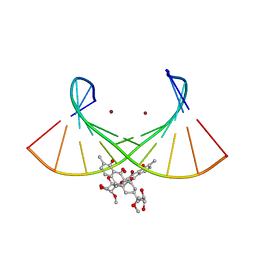 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
5YZE
 
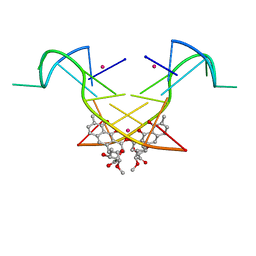 | | Crystal structure of the [Co2+-(chromomycin A3)2]-d(CCG)3 complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Chen, Y.W, Wu, P.C, Satange, R.B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Int J Mol Sci, 19, 2018
|
|
7Y2P
 
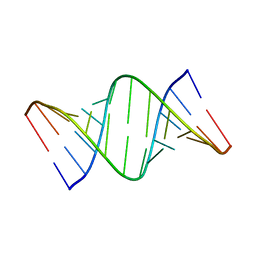 | |
5XJW
 
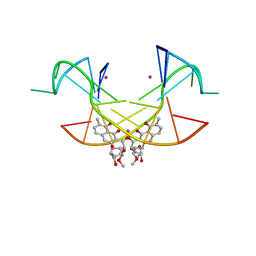 | | Crystal Structure of the [Co2+-(Chromomycin A3)2]-CCG repeats Complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Tseng, W.H, Wu, P.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal Structure of the [Co2+-(Chromomycin A3)2]-CCG repeats Complex
To be published
|
|
7Y2B
 
 | |
