1GNN
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASN (V82N) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1GNM
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASP (V82D) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
3I7E
 
 | | Co-crystal structure of HIV-1 protease bound to a mutant resistant inhibitor UIC-98038 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, HIV-1 protease | | Authors: | Hong, L, Tang, J, Ghosh, A. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
1CJY
 
 | | HUMAN CYTOSOLIC PHOSPHOLIPASE A2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, PROTEIN (CYTOSOLIC PHOSPHOLIPASE A2) | | Authors: | Dessen, A, Tang, J, Schmidt, H, Stahl, M, Clark, J.D, Seehra, J, Somers, W.S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
4GID
 
 | | Structure of beta-secretase complexed with inhibitor | | Descriptor: | Beta-secretase 1, L-PROLINAMIDE, N-[(2S)-1-({(2S,3R)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-3-phenylpropan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A, Tang, J, Venkateswara, R.K, Yadav, N, Anderson, D, Gavande, N, Huang, X, Terzyan, S. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
J.Med.Chem., 55, 2012
|
|
5D1V
 
 | |
1YWN
 
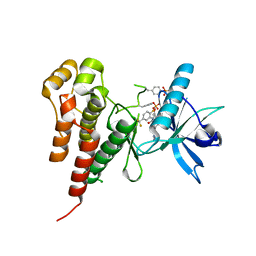 | | Vegfr2 in complex with a novel 4-amino-furo[2,3-d]pyrimidine | | Descriptor: | N-{4-[4-AMINO-6-(4-METHOXYPHENYL)FURO[2,3-D]PYRIMIDIN-5-YL]PHENYL}-N'-[2-FLUORO-5-(TRIFLUOROMETHYL)PHENYL]UREA, Vascular endothelial growth factor receptor 2 | | Authors: | Miyazaki, Y, Matsunaga, S, Tang, J, Maeda, Y, Nakano, M, Philippe, R.J, Shibahara, M, Liu, W, Sato, H, Wang, L, Nolte, R.T. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Novel 4-amino-furo[2,3-d]pyrimidines as Tie-2 and VEGFR2 dual inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1F42
 
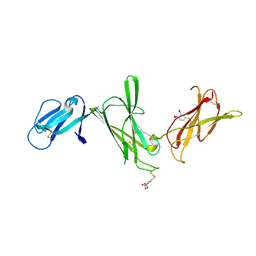 | | THE P40 DOMAIN OF HUMAN INTERLEUKIN-12 | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
1F45
 
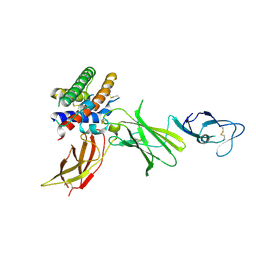 | | HUMAN INTERLEUKIN-12 | | Descriptor: | INTERLEUKIN-12 ALPHA CHAIN, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
3RE9
 
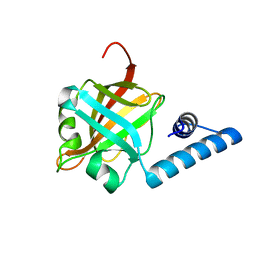 | | Crystal structure of sortaseC1 from Streptococcus suis | | Descriptor: | Sortase-like protein | | Authors: | Lu, G, Qi, J, Gao, F, Yan, J, Tang, J, Gao, G.F. | | Deposit date: | 2011-04-04 | | Release date: | 2011-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel "open-form" structure of sortaseC from Streptococcus suis.
Proteins, 79, 2011
|
|
3QK0
 
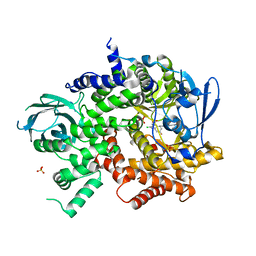 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QJZ
 
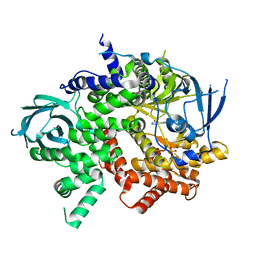 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
1XN2
 
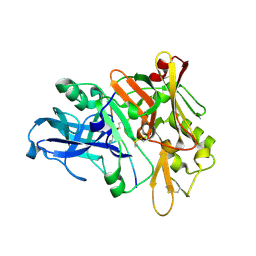 | | New substrate binding pockets for beta-secretase. | | Descriptor: | Beta-secretase 1, OM03-4 | | Authors: | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
1XN3
 
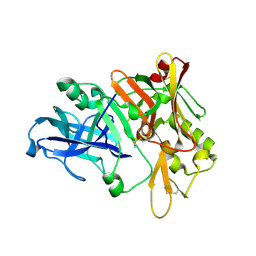 | | Crystal structure of Beta-secretase bound to a long inhibitor with additional upstream residues. | | Descriptor: | Beta-secretase 1, Peptidic inhibitor | | Authors: | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
4RA2
 
 | | PP2Ca | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2Ca
SCI REP, 2015
|
|
4RAG
 
 | | CRYSTAL STRUCTURE of PPC2A-D38K | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
4RAF
 
 | | Crystal structure of PP2Ca-D38A | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
1XS7
 
 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2M2R
 
 | | Solution structure of MCh-2: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-2 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
5DQC
 
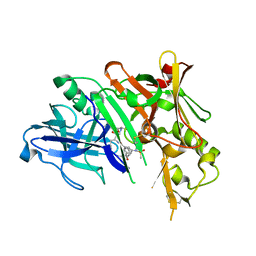 | | Co-crystal of BACE1 with compound 0211 | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-3-hydroxy-4-({(2S,3S)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A.K, Bhavanam, S.R, Yen, T.-C, Cardenas, E.L, Rao, K.V, Downs, D, Huang, X, Tang, J, Mescar, A.D. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-17 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.4651 Å) | | Cite: | Design of Potent and Highly Selective Inhibitors for Human beta-Secretase 2 (Memapsin 1), a Target for Type 2 Diabetes.
Chem Sci, 7, 2016
|
|
1SGZ
 
 | |
1OXZ
 
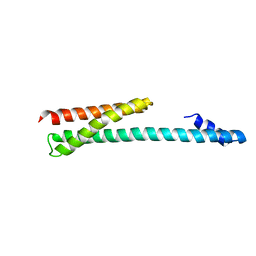 | | Crystal Structure of the Human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Zhu, G, Zhai, P, He, X, Terzyan, S, Zhang, R, Joachimiak, A, Tang, J, Zhang, X.C. | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human GGA1 GAT Domain
Biochemistry, 42, 2003
|
|
1Z52
 
 | | Proaerolysin Mutant W373L | | Descriptor: | Aerolysin | | Authors: | Parker, M.W, Feil, S.C, Tang, J.W. | | Deposit date: | 2005-03-16 | | Release date: | 2006-03-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Proaerolysin at 2.3 A Resolution and Structural Analyses of Single-site Mutants as a Basis for Understanding Membrane Insertion of the Toxin
To be Published
|
|
2G94
 
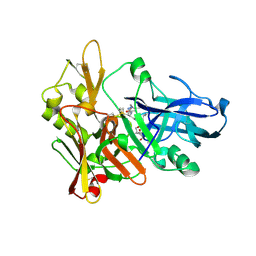 | | Crystal structure of beta-secretase bound to a potent and highly selective inhibitor. | | Descriptor: | Beta-secretase 1, N~2~-[(2R,4S,5S)-5-{[N-{[(3,5-DIMETHYL-1H-PYRAZOL-1-YL)METHOXY]CARBONYL}-3-(METHYLSULFONYL)-L-ALANYL]AMINO}-4-HYDROXY-2,7-DIMETHYLOCTANOYL]-N-ISOBUTYL-L-VALINAMIDE | | Authors: | Hong, L, Ghosh, A, Tang, J. | | Deposit date: | 2006-03-05 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design, synthesis and X-ray structure of protein-ligand complexes: important insight into selectivity of memapsin 2 (beta-secretase) inhibitors.
J.Am.Chem.Soc., 128, 2006
|
|
