2Q9N
 
 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1S,4R,7AR)-4-BUTOXY-1-[(1R)-1-FORMYLPROPYL]-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
7BR5
 
 | | Lysozyme-sugar complex in H2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3FVR
 
 | |
3FVT
 
 | |
3FYT
 
 | |
3FYU
 
 | |
7U1J
 
 | |
3FCY
 
 | |
2ZY6
 
 | | Crystal structure of a truncated tRNA, TPHE39A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Tanaka, I, Yao, M, Tanaka, Y, Kitago, Y, Ymagata, S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand
Rna, 15, 2009
|
|
7DER
 
 | | Lysozyme alone in H2O | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DEQ
 
 | | Lysozyme-sugar complex in D2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2Q9M
 
 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
1VE0
 
 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | Descriptor: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | Authors: | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
4WDR
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WDQ
 
 | | Crystal structure of haloalkane dehalogenase LinB32 mutant (L177W) from Sphingobium japonicum UT26 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, MAGNESIUM ION | | Authors: | Degtjarik, O, Rezacova, P, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WCV
 
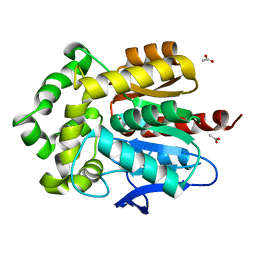 | | Haloalkane dehalogenase DhaA mutant from Rhodococcus rhodochrous (T148L+G171Q+A172V+C176G) | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Holubeva, T, Prudnikova, T, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2014-09-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Balancing the stability-activity trade-off by fine-tuning dehalogenase access tunnels
Chemcatchem, 2015
|
|
4F60
 
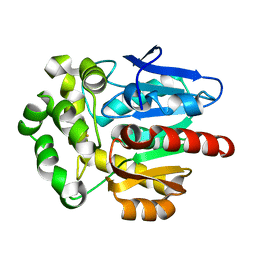 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (T148L, G171Q, A172V, C176F). | | Descriptor: | FLUORIDE ION, Haloalkane dehalogenase | | Authors: | Plevaka, M, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4F5Z
 
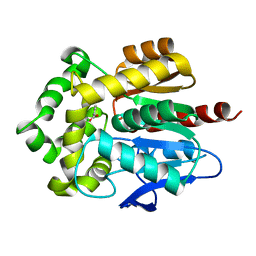 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (L95V, A172V). | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Kulik, D, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1VCX
 
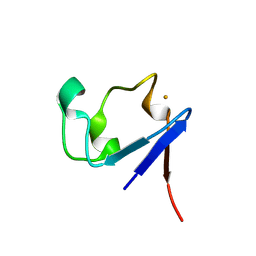 | | Neutron Crystal Structure of the Wild Type Rubredoxin from Pyrococcus Furiosus at 1.5A Resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Kurihara, K, Tanaka, I, Chatake, T, Adams, M.W.W, Jenney Jr, F.E, Moiseeva, N, Bau, R, Niimura, N. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Neutron crystallographic study on rubredoxin from Pyrococcus furiosus by BIX-3, a single-crystal diffractometer for biomacromolecules
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VGJ
 
 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
4F3F
 
 | | Crystal Structure of Msln7-64 MORAb-009 FAB complex | | Descriptor: | MORAb-009 Fab heavy chain, MORAb-009 Fab light chain, Mesothelin | | Authors: | Xia, D, Pastan, I, Ma, J, Tang, W.K, Esser, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
J.Biol.Chem., 287, 2012
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FJ1
 
 | | Crystal Structure of the ternary complex between a fungal 17beta-hydroxysteroid dehydrogenase (Holo form) and genistein | | Descriptor: | 17beta-hydroxysteroid dehydrogenase, DIMETHYL SULFOXIDE, GENISTEIN, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I. | | Deposit date: | 2012-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
2F7L
 
 | | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase | | Descriptor: | 455aa long hypothetical phospho-sugar mutase, SULFATE ION | | Authors: | Kawamura, T, Sakai, N, Akutsu, J, Zhang, Z, Watanabe, N, Kawarabayashi, Y, Tanaka, I. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase
To be Published
|
|
3U2J
 
 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
