5GSR
 
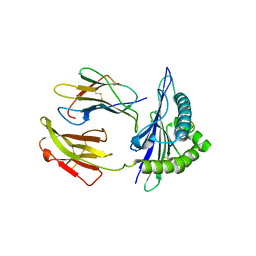 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | 分子名称: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | 登録日 | 2016-08-17 | | 公開日 | 2017-04-26 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
6BF8
 
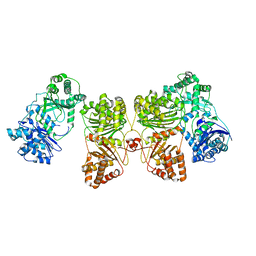 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Y
 
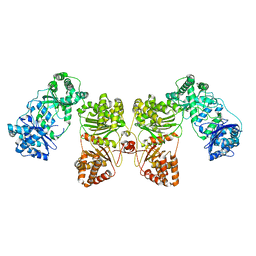 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BFC
 
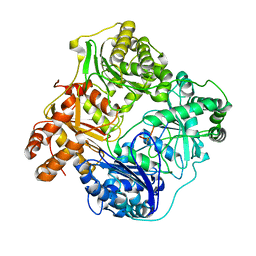 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF6
 
 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-09-22 | | 公開日 | 2017-11-22 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | 分子名称: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2018-01-10 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
3N57
 
 | |
5GWY
 
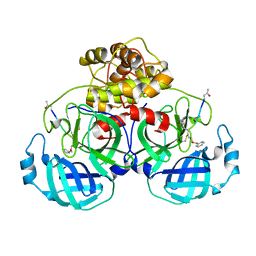 | | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design | | 分子名称: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, main protease | | 著者 | Wang, F, Chen, C, Tan, W, Yang, K, Yang, H. | | 登録日 | 2016-09-14 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.852 Å) | | 主引用文献 | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design.
Sci Rep, 6, 2016
|
|
5V56
 
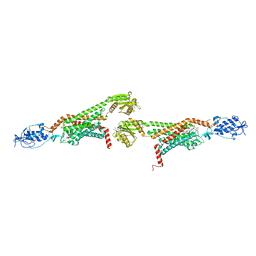 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | 著者 | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | 登録日 | 2017-03-13 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
5V57
 
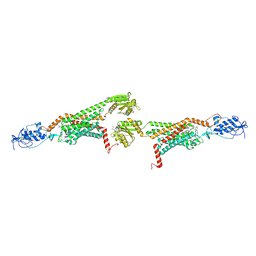 | | 3.0A SYN structure of the multi-domain human smoothened receptor in complex with TC114 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | 著者 | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | 登録日 | 2017-03-13 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
5XIV
 
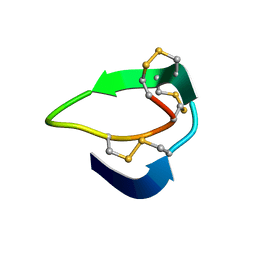 | | Beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba | | 分子名称: | beta-ginkgotide, beta-gB1 | | 著者 | Wong, K.H, Tan, W.L, Xiao, T, Tam, J.P. | | 登録日 | 2017-04-27 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba.
Sci Rep, 7, 2017
|
|
5B7C
 
 | |
5BT2
 
 | | MeCP2 MBD domain (A140V) in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*TP*AP*GP*AP*AP*GP*AP*AP*TP*TP*CP*(5CM)P*GP*TP*TP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*GP*AP*AP*(5CM)P*GP*GP*AP*AP*TP*TP*CP*TP*TP*CP*TP*A)-3'), Methyl-CpG-binding protein 2 | | 著者 | Ho, K.L, Chia, J.Y, Tan, W.S, Ng, C.L, Hu, N.J, Foo, H.L. | | 登録日 | 2015-06-02 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence.
Sci Rep, 6, 2016
|
|
3SNS
 
 | | Crystal structure of the C-terminal domain of Escherichia coli lipoprotein BamC | | 分子名称: | CHLORIDE ION, Lipoprotein 34 | | 著者 | Kim, K.H, Aulakh, S, Tan, W, Paetzel, M. | | 登録日 | 2011-06-29 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4MOD
 
 | | Structure of the MERS-CoV fusion core | | 分子名称: | HR1 of S protein, LINKER, HR2 of S protein | | 著者 | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | 登録日 | 2013-09-12 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
2YSO
 
 | | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog | | 分子名称: | ZINC ION, Zinc finger protein 95 homolog | | 著者 | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-03 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog
To be Published
|
|
2YSP
 
 | | Solution structure of the C2H2 type zinc finger (region 507-539) of human Zinc finger protein 224 | | 分子名称: | ZINC ION, Zinc finger protein 224 | | 著者 | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-03 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C2H2 type zinc finger (region 507-539)of human Zinc finger protein 224
To be Published
|
|
8GJM
 
 | | 17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | 登録日 | 2023-03-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8GJN
 
 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | 著者 | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | 登録日 | 2023-03-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
3KBX
 
 | |
3N56
 
 | |
8H0J
 
 | | Annexin A5 mutant | | 分子名称: | Annexin A5, CALCIUM ION | | 著者 | Hua, Z.C, Tang, W. | | 登録日 | 2022-09-29 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | structure dissection of the membrane aggregation mechanism induced by Annexin A5 mutation
To Be Published
|
|
