4V7L
 
 | | The structures of viomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3K0E
 
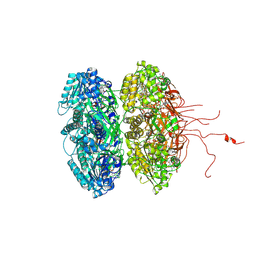 | | Crystal structure of the phosphorylation-site mutant T426N of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
4V7M
 
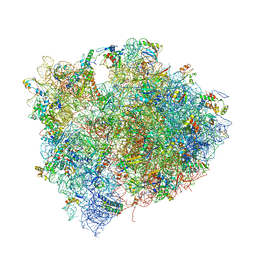 | | The structures of Capreomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6M87
 
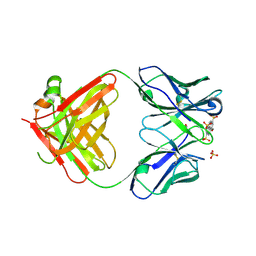 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6J13
 
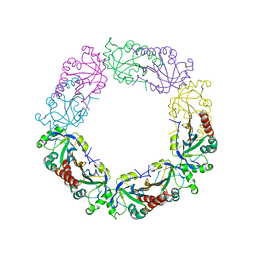 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
4X6Z
 
 | | Yeast 20S proteasome in complex with PR-VI modulator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rostankowski, R, Witkowska, J, Borek, D, Otwinowski, Z, Jankowska, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures revealed the common place of binding of low-molecular
mass activators with the 20S proteasome
To Be Published
|
|
5FJS
 
 | | Bacterial beta-glucosidase reveals the structural and functional basis of genetic defects in human glucocerebrosidase 2 (GBA2) | | Descriptor: | CALCIUM ION, GLUCOSYLCERAMIDASE | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Breen, I, Mutoha, R, Sansenya, S, Hua, Y, Tankrathok, A, Wu, L, Songsiriritthigul, C, Tanaka, H, Williams, S.J, Davies, G.J, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-10-12 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bacterial Beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (Gba2)
Acs Chem.Biol., 11, 2016
|
|
4V6A
 
 | | Structure of EF-P bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome.
Science, 325, 2009
|
|
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
3K09
 
 | | Crystal structure of the phosphorylation-site mutant S431D of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3JZM
 
 | | Crystal structure of the phosphorylation-site mutant T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-23 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0A
 
 | | Crystal structure of the phosphorylation-site mutant S431A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0F
 
 | | Crystal structure of the phosphorylation-site double mutant T426A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0C
 
 | | Crystal structure of the phosphorylation-site double mutant S431A/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
5J74
 
 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
5J75
 
 | | Fluorogen Activating Protein AM2.2 in complex with ML342 | | Descriptor: | N,4-dimethyl-N-{2-oxo-2-[4-(pyridin-2-yl)piperazin-1-yl]ethyl}benzene-1-sulfonamide, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
8EDF
 
 | |
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECV
 
 | | Bovine Fab 2F12 | | Descriptor: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5BX4
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with Glucoimidazole | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLUCOIMIDAZOLE, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
7R1Y
 
 | |
6OPA
 
 | |
5BX3
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
